Abstract
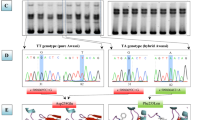
Some mutations in BMP15 gene (Bone morphogenetic protein 15) have been known to be associated with prolificacy in various breeds, which can regulate follicular development, granulosa cell proliferation, and cumulus differentiation, etc., and plays a crucial role in the regulation of female animal fertility. Polymorphism of BMP15 gene and its reproductive traits of Bamei pigs was studied by using DNA sequencing. Bioinformatics tools were used to find QTLs that affected the reproductive traits of Bamei pigs, so as to provide a basis for molecular marker-assisted selection. Which Bamei pigs were served as research objects, the polymorphic locus of BMP15 gene were detected by using DNA direct sequencing and analyze genotype. Sus scrofa BMP15 protein has a lot of similar structures with human and other mammals in the primary, secondary, and tertiary structures; and closely related to cattle in terms of genetic evolution. There were three SNPs sites found in BMP15 gene of the test group, which g.5969C > T, g.6543C > T, g.6653A > G, the results of Chi-square test indicated that three SNPs loci fitted in Hardy–Weinberg equilibrium (p > 0.05). Type1 and Type3 mutations were beneficial to increasing the litter size in Qinghai Bamei pigs, and the litter size by homozygous TT of Type1 mutation was significantly higher than wild homozygous CC and heterozygous CT (P < 0.05). The results of the present study indicated the potential of the observed SNP of BMP15 for further exploitation in marker-assisted selection to improve reproduction efficiency of Bamei pigs.






Similar content being viewed by others
REFERENCES
Nicol, L., Bishop, S.C., Pong-Wong, R., et al., Homozygosity for a single base-pair mutation in the oocyte-specific GDF9 gene results in sterility in Thoka sheep, Reproduction, 2009, vol. 138, no. 6, pp. 921—933. https://doi.org/10.1530/REP-09-0193
Hanrahan, J.P., Gregan, S.M., Mulsant, P., et al., Mutations in the genes for oocyte-derived growth factors GDF9 and BMP15 are associated with both increased ovulation rate and sterility in Cambridge and Belclare sheep (Ovis aries), Biol. Reprod., 2004, vol. 70, no. 4, pp. 900—909. https://doi.org/10.1095/biolreprod.103.023093
Shimasaki, S., Moore, R.K., Otsuka, F., and Erickson, G.F., The bone morphogenetic protein system in mammalian reproduction, Endocr. Rev., 2004, vol. 25, no. 1, pp. 72—101. https://doi.org/10.1210/er.2003-00007
Knight, P.G. and Glister, C., TGF-beta superfamily members and ovarian follicle development, Reproduction, 2006, vol. 132, no. 2, pp. 191—206. https://doi.org/10.1530/rep.1.01074
Otsuka, F., Yao, Z., Lee, T., et al., Bone morphogenetic protein-15: identification of target cells and biological functions, J. Biol. Chem., 2000, vol. 275, no. 50, pp. 39523—39528. https://doi.org/10.1074/jbc.M007428200
Kona, S.S., Praveen Chakravarthi, V., Siva Kumar, A.V., et al., Quantitative expression patterns of GDF9 and BMP15 genes in sheep ovarian follicles grown in vivo or cultured in vitro, Theriogenology, 2016, vol. 85, no. 2, pp. 315—322. https://doi.org/10.1016/j.theriogenelogy.2015.09.022
Paz, E., Quiñones, J., Bravo, S., et al., Genotyping of BMPR1B, BMP15 and GDF9 genes in Chilean sheep breeds and association with prolificacy, Anim. Genet., 2015, vol. 46, no. 1, pp. 98—99. https://doi.org/10.1111/age12254
Paulini, F. and Melo, E.O., The role of oocyte-secreted factors GDF9 and BMP15 in follicular development and oogenesis, Reprod. Domest. Anim., 2011, vol. 46, no. 2, pp. 354—361. https://doi.org/10.1111/j.1439-0531.2010.01739.x
Galloway, S.M., McNatty, K.P., Cambridge, L.M., et al., Mutations in an oocyte-derived growth factor gene (BMP15) cause increased ovulation rate and infertility in a dosage-sensitive manner, Nat. Genet., 2000, vol. 25, no. 3, pp. 279—283. https://doi.org/10.1038/77033
Vinet, A., Drouilhet, L., Bodin, L., et al., Genetic control of multiple births in low ovulating mammalian species, Mamm. Genome, 2012, vol. 23, nos. 11—12, pp. 727—740. https://doi.org/10.1007/s00335-012-9412-4
Clelland, E., Kohli, G., Campbell, R.K., et al., Bone morphogenetic protein-15 in the zebrafish ovary: complementary deoxyribonucleic acid cloning, genomic organization, tissue distribution, and role in oocyte maturation, Endocrinology, 2006, vol. 147, no. 1, pp. 201—209.https://doi.org/10.1210/en.2005-1017
Elis, S., Dupont, J., Couty, I., et al., Expression and biological effects of bone morphogenetic protein-15 in the hen ovary, J. Endocrinol., 2007, vol. 194, no. 3, pp. 485—497.https://doi.org/10.1677/JOE-07-0143
Huang, H.Y., Liang, Z., Li, S.F., et al., Polymorphism identification in BMP15 and GDF9 genes and their association with egg production in chickens, Br. Poult. Sci., 2015, vol. 56, no. 3, pp. 277—283. https://doi.org/10.1080/00071668.2015.1019829
Pramod, R.K., Sharma, S.K., Singhi, A., et al., Differential ovarian morphometry and follicular expression of BMP15, GDF9 and BMPR1B influence the prolificacy in goat, Reprod. Domest. Anim., 2013, vol. 48, no. 5, pp. 803—809.https://doi.org/10.1111/rda.12165
Monestier, O., Servin, B., Auclair, S., et al., Evolutionary origin of bone morphogenetic protein 15 and growth and differentiation factor 9 and differential selective pressure between mono- and polyovulating species, Biol. Reprod., 2014, vol. 91, no. 4, p. 83.https://doi.org/10.1095/biolreprod.114.119735
Hall, B.G., Building phylogenetic trees from molecular data with MEGA, Mol. Biol. Evol., 2013, vol. 30, no. 5, pp. 1229—1235.https://doi.org/10.1093/molbev/mst012
Bailey, T.L., Boden, M., Buske, F.A., et al., MEME SUITE: tools for motif discovery and searching, Nucleic Acids Res., 2009, vol. 37, pp. W202—W208. https://doi.org/10.1093/nar/gkp335
Chen, C., Rui, X., Hao, C., et al., TBtools, a toolkit for biologists integrating various HTS-data handling tools with a user-friendly interface, bioRxiv, 2018, p. 289660.
von Mering, C., Jensen, L.J., Snel, B., et al., STRING: known and predicted protein—protein associations, integrated and transferred across organisms, Nucleic Acids Res., 2005, vol. 33, pp. D433—D437.https://doi.org/10.1093/nar/gki005
Shannon, P., Markiel, A., Ozier, O., et al., Cytoscape: a software environment for integrated models of biomolecular interaction networks, Genome Res., 2003, vol. 13, no. 11, pp. 2498—2504. https://doi.org/10.1101/gr.1239303
Shi, Y.Y. and He, L., SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci, Cell Res., 2005, vol. 15, no. 2, pp. 97—98. https://doi.org/10.1038/sj.cr.7290272
Liu, X., Ma, L., Wang, M., et al., Two indel variants of prolactin receptor (PRLR) gene are associated with growth traits in goat, Anim. Biotechnol., 2019, pp. 1—10. https://doi.org/10.1080/10495398.2019/1594863
Wang, X., Yang, Q., Wang, K., et al., A novel 12-bp indel polymorphism within the GDF9 gene is significantly associated with litter size and growth traits in goats, Anim. Genet., 2017, vol. 48, no. 6, pp. 735—736. https://doi.org/10.1111/age.12617
Yang, Q., Yan, H., Li, J., et al., A novel 14-bp duplicated deletion within goat GHR gene is significantly associated with growth traits and litter size, Anim. Genet., 2017, vol. 48, no. 4, pp. 499—500. https://doi.org/10.1111/age.12551
Zhao, C., Gui, L., Li, Y., et al., Associations between allelic polymorphism of the BMP binding endothelial regulator and phenotypic variation of cattle, Mol. Cell Probes, 2015, vol. 29, no. 6, pp. 358—364. https://doi.org/10.1016/j.mcp.2015.09.007
Nagdy, H., Mahmoud, K.G.M., Kandiel, M.M.M., et al., PCR-RFLP of bone morphogenetic protein 15 (BMP15/FecX) gene as a candidate for prolificacy in sheep, Int. J. Vet. Sci. Med., 2018, vol. 6, suppl., pp. S68—S72. https://doi.org/10.1016/j.ijvsm.2018.01.001
Chantepie, L., Bodin, L., Sarry, J., et al., Genome-wide identification of a regulatory mutation in BMP15 controlling prolificacy in sheep, Front. Genet., 2020, vol. 11, p. 585. https://doi.org/10.3389/fgene.2020.00585
Monteagudo, L.V., Ponz, R., Tejedor, M.T., et al., A 17 bp deletion in the bone morphogenetic protein 15 (BMP15) gene is associated to increased prolificacy in the Rasa Aragonesa sheep breed, Anim. Reprod. Sci., 2009, vol. 110, nos. 1–2, pp. 139—146. https://doi.org/10.1016/j.anireprosci.2008.01.005
Zamani, P., Nadri, S., Saffaripour, R., et al., A new mutation in exon 2 of the bone morphogenetic protein 15 gene is associated with increase in prolificacy of Mehraban and Lori sheep, Trop. Anim. Health Prod., 2015, vol. 47, no. 5, pp. 855—860. https://doi.org/10.1007/s11250-015-0799-2
Jia, J., Chen, Q., Gui, L., et al., Association of polymorphisms in bone morphogenetic protein receptor-1B gene exon-9 with litter size in Dorset, Mongolian, and Small Tail Han ewes, Asian-Australas. J. Anim. Sci., 2019, vol. 32, no. 7, pp. 949—955. https://doi.org/10.5713/ajas.18.0541
Lassoued, N., Benkhlil, Z., Woloszyn, F., et al., FecX (Bar) a novel BMP15 mutation responsible for prolificacy and female sterility in Tunisian Barbarine sheep, BMC Genet., 2017, vol. 18, no. 1, p. 43. https://doi.org/10.1186/s12863-017-0510-x
Demars, J., Fabre, S., Sarry, J., et al., Genome-wide association studies identify two novel BMP15 mutations responsible for an atypical hyperprolificacy phenotype in sheep, PLoS Genet., 2013, vol. 9, no. 4. e1003482 https://doi.org/10.1371/journal.pgen1003482
Patil, N., Berno, A.J., Hinds, D.A., et al., Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21, Science, 2001, vol. 294, no. 5547, pp. 1719—1723. https://doi.org/10.1371/journal.pgen.1003482
ACKNOWLEDGMENTS
The authors are grateful to the staff at the original breeding farm of the Huzhu Bamei pigs in Qinghai for collecting samples.
Funding
This work was supported by the Scientific and Technological Achievements Transformation Project of Qinghai Provincial Department of Science and Technology (2019-NK-102).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest. The authors declare that they have no conflicts of interest.
Statement on the welfare of animals. All experimental procedures were approved by the Review Committee for the Use of Qinghai University. Animal experimentation, including sample collection, was performed in agreement with the ethics commission’s guidelines. The use of experimental animals was permitted by local animal welfare laws, guidelines, and policies.
Rights and permissions
About this article
Cite this article
Shen, W., Wang, L., Ma, Y. et al. Association between BMP15 Gene Polymorphisms of Growth Traits and Litter Size in Qinghai Bamei Pigs. Russ J Genet 58, 997–1006 (2022). https://doi.org/10.1134/S1022795422080075
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795422080075