Abstract
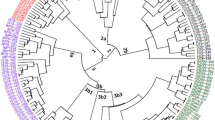
This study was carried out to estimate the genetic diversity between cultivars of alfalfa using SRAP marker system (sequence-related amplified polymorphism). We tested 25 combinations of SRAP markers and revealed that seven of them were informative for DNA polymorphism identification. These combinations generated 129 PCR products with the percentage of polymorphic bands per pair ranging from 21 to 50%. Specific amplification products were identified for 14 cultivars for genetic certification.



Similar content being viewed by others
REFERENCES
Piskovatskii, Yu.M., Kosolapov, V.M., Mikhalev, V.E., et al., Agrotekhnika vozdelyvaniya sortov lyutserny selektsii VNII kormov im. V.R. Vil’yamsa na semennye i kormovye tseli (Agronomic Technics for Cultivation of Alfalfa Cultivars Bred by the All-Russian Williams Federal Science Center for Fodder Production and Agroecology for Seed and Forage Purpose), Moscow: Rossiyskii Tsentr Sel’skokhozyaistvennogo Konsul’tirovaniya, 2008, pp. 3—15.
Veronesi, F., Brummer, E.C., and Huyghe, C., Alfalfa, in Fodder Crops and Amenity Grasses, New York: Springer-Verlag, 2010, pp. 395—437.
Chesnokov, Yu.V. and Kosolapov, V.M., Geneticheskie resursy rastenii i uskorenie selektsionnogo protsessa (Plant Genetic Resources and Acceleration of the Breeding Process), Moscow: Ugreshskaya Tipografiya, 2016.
Piskovatskii, Yu.M., Lomova, M.G., Solozhentseva, L.F., et al., Creation of promising alfalfa material with high seed productivity, Nauchnoe obespechenie kormoproizvodstva i ego rol’ v sel’skom khozyaistve, ekonomike, ekologii i ratsional’nom prirodopol’zovanii Rossii (Scientific Support of Fodder Production and its Role in Agriculture, Economy, Ecology and Rational Use of Natural Resources in Russia) (Proc. Int. Theor. Pract. Conf., in Memoriam of Acad. A.A. Zhuchenko, Lobnya, June 19—20, 2013), Moscow: Ugreshskaya Tipografiya, 2013, pp. 97—103.
Khlestkina, E.K., Molecular markers in genetic studies and breeding, Russ. J. Genet.: Appl. Res., 2013, vol. 4, no. 3, pp. 236—244.
Khlestkina, E.K., Molecular methods for analyzing the structure—function organization of genes and genomes in higher plants, Russ. J. Genet.: Appl. Res., 2011, vol. 15, pp. 757—768.
Li, G. and Quiros, C.F., Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: its application to mapping and gene tagging in Brassica, Theor. Appl. Genet., 2001, vol. 103, nos. 2—3, pp. 455—461. https://doi.org/10.1007/s001220100570
Osman, G., Suleyman, K., and Kazim, A., Diversity and relationships among Turkish okra germplasm by SRAP and phenotypic marker polymorphism, Biologia (Bratislava), 2007, vol. 62, no. 1, pp. 41—45. https://doi.org/10.2478/s11756-007-0010-y
Lin, Z., Zhang, X., Nie, Y., et al., Construction of a genetic linkage map for cotton based on SRAP, Chin. Sci. Bull., 2003, vol. 48, no. 19, pp. 2064—2068. https://doi.org/10.1360/03wc0193
Luo, Y.X., Du, D.Z., Fu, G., et al., Inheritance of leaf color and sequence-related amplified polymorphic (SRAP) molecular markers linked to the leaf color gene in Brassica juncea, Afr. J. Biotechnol., 2011, vol. 10, no. 66, pp. 14724—14730. https://doi.org/10.5897/AJB11.1096
Hao, Q., Liu, Z.A., Shu, Q.Y., et al., Studies on Paeonia cultivars and hybrids identification based on SRAP analysis, Hereditas, 2008, vol. 145, no. 1, pp. 38—47. https://doi.org/10.1111/j.0018-0661.2008.2013.x
Guo, D.L. and Luo, Z.R., Genetic relationships of some PCNA persimmons (Diospyros kaki Thumb.) from China and Japan revealed by SRAP analysis, Genet. Resour. Crop. Evol., 2006, vol. 53, no. 8, pp. 1597—1603. https://doi.org/10.1007/s10722-005-8717-5
Dellaporta, S.L., Wood, J., and Hicks, J.B., A plant DNA minipreparation: version II, Plant Mol. Biol. Rep., 1983, vol. 1, no. 4, pp. 19—21.
Robarts, D.W.H. and Wolfe, A.D., Sequence-related amplified polymorphism (SRAP) markers: a potential resource for studies in plant molecular biology, Appl. Plant Sci., 2014, vol. 2, no. 7, p. 1400017. https://doi.org/10.3732/apps.1400017
Aneja, B., Yadav, N.R., Chawla, V., and Yadav, R.C., Sequence-related amplified polymorphism (SRAP) molecular marker system and its applications in crop improvement, Mol. Breed., 2012, vol. 30, no. 4, pp. 1635—1648. https://doi.org/10.1007/s11032-012-9747-2
Vandemark, J.G., Ariss, J.J. Bauchan, G.A., et al., Estimating genetic relationships among historical sources of alfalfa germplasm and selected cultivars with sequence related amplified polymorphisms, Euphytica, 2006, vol. 152, no. 1, pp. 9—16. https://doi.org/10.1007/s10681-006-9167-7
Rhouma, H.B., Taski-Ajdukovic, K., Zitouna, N., et al., Assessment of the genetic variation in alfalfa genotypes using SRAP markers for breeding purposes, Chil. J. Agric. Res., 2017, vol. 77, no. 4, pp. 332—339. https://doi.org/10.4067/S0718-58392017000400332
Al-Faifi, S., Migdadi, H., Al-Doss, A., et al., Morphological and molecular genetic variability analyses of Saudi lucerne (Medicago sativa L.), Crop Pasture Sci., 2013, vol. 64, no. 2, pp. 137—146. https://doi.org/10.1071/CP12271
Talebi, M.B., Hajiahmadi, Z., and Rahimmalek, M., Genetic diversity and population structure of four Iranian alfalfa populations revealed by sequence-related amplified polymorphism (SRAP) markers, J. Crop. Sci. Biotech., 2011, vol. 14, no. 3, pp. 173—178. https://doi.org/10.1007/s12892-011-0030-6
Yuan, Q., Gao, J., Gui, Z., et al., Genetic relationships among alfalfa germplasms resistant to common leaf spot and selected Chinese cultivars assessed by sequence-related amplified polymorphism (SRAP) markers, Afr. J. Biotech., 2011, vol. 10, no. 59, pp. 12527—12534. https://doi.org/10.5897/AJB11.901
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
Rights and permissions
About this article
Cite this article
Shamustakimova, A.O., Mavlyutov, Y.M. & Klimenko, I.A. Application of SRAP Markers for DNA Identification of Russian Alfalfa Cultivars. Russ J Genet 57, 540–547 (2021). https://doi.org/10.1134/S1022795421050112
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795421050112