Abstract
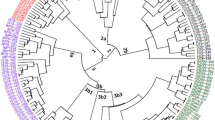
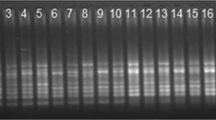
Characterization of germplasm is an important pathway between the conservation and utilization of genetic resources in various breeding programs. Genetic variability and genetic relationships were accessed among the 42 genotypes of Garcinia L. of Upper Assam, using the ISSR-DNA markers. A total of 42 random decamer oligonucleotides of PKBT and UBC series were examined to generate the ISSR profiling, out of which 16 primers produced reproducible and scorable bands. A total of 142 amplicons was obtained, with 91.56% of polymorphic bands. Monomorphic band percentage was calculated and found to be 8.48% with 0.65 of PIC value and EMR, MI values were calculated and found to be 7.90 and 5.52, respectively. Genetic similarity matrix, based on Jaccard’s coefficient ranged from 0.30 to 0.97 indicating to be least. A dendrogram constructed by the unweighted pair group method with arithmetic average formed four distinct clusters. ITS1 regions were found to be efficient barcode for the assessment of phylogenetic relationships between Garcinia genotypes. The present investigation recommends the usefulness of ISSR technology for the study of genetic similarity among different genotypes of Garcinia L.






Similar content being viewed by others
References
Cox JEK (1976) Garcinia mangostana-Mangosteen In R. J. Garner and S. Ahmed Chaudhari (Eds.) The propagation of tropical fruit trees. Horticultural Review East Malling: Commonwealth Bureau of Horticulture and Plantation Crops. 4; 361–375.
Stevens PF (2007) Clusiaceae-Guttiferae Flowering Plants Eudicots vol.1 pp.48–66 Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-540-32219-1_10
Burkill IH (1935) A dictionary of the economic products of the Malay Peninsula A Dictionary of the Economic Products of the Malay Peninsula. vol.2; 2444
Hooker, JD (1872–1897) The Flora of British India Vol.1 Reeve and Co, London
Kanjilal UN, Kanjilal PC, Das A, De RN, Bor NL (1934–40) Flora of Assam Vol.1–5. Government Press, Shillong
Rabbani MA, Masood MS, Shinwari ZK, Shinozaki KY (2010) Genetic analysis of basmati and non-basmati Pakistani rice cultivars using microsatellite markers. Pak J Bot. 42(4):2551–2564
Kumar P, Gambhir G, Gaur A, Srivastava DK (2015) Molecular analysis of genetic stability in in vitro regenerated plants of broccoli. Curr Sci. 109(8):1470–1475
Gupta VK, Castro SL, Over TM (1996) On scaling exponents of spatial peak flows from rainfall and river network geometry. J Hydrol 187:81–104. https://doi.org/10.1016/S0022-1694(96)03088-0
Doyle JJ, Doyle JJ (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15. https://doi.org/10.2307/2419362
Poczai P, Varga I, Laos M, Cseh A, Bell N, Valkonen JP, Hyvönen J (2013) Advances in plant gene-targeted and functional markers: a review. Plant Methods 9(1):6
Jugran AK, Bhatt ID, Mondal S, Rawal RS, Nandi, SK (2015) Genetic diversity assessment of Valeriana jatamansi Jones using microsatellites markers. Curr Sci. https://doi.org/10.18520/cs/v109/i7/1273-1282
Gerbi, S. A. (1985) Evolution of ribosomal DNA Molecular evolutionary genetics Plenum Press. 419–517.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410. https://doi.org/10.1016/S0022-2836(05)80362
Jaccard P (1908) Nouvelles Recherches Sur la distribution floral Bulletin de la Société vaudoise des sciences naturelles 44:223–270
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1686077
Gilbert JE, Lewis RV, Wilkinson MJ, Caligari PDS (1999) Developing an appropriate strategy to assess genetic variability in plant germplasm collections. Theor Appl Genet 98:1125–1131. https://doi.org/10.1007/s001220051176
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599. https://doi.org/10.1093/molbev/msaa014
Baldwin BG, Sanderson MJ, Porter JM, Wojciechowski MF, Campbell CS, Donoghue MJ (1995) The ITS region of nuclear ribosomal DNA: a valuable source of evidence on angiosperm phylogeny. Ann Missouri Bot Gard 82:247–277. https://doi.org/10.2307/2399880
Stewart MA, Hall LMC, Maden BEH (1983) Multiple heterogeneities in the transcribed spacers of ribosomal DNA from Xenopus laevis. Nucleic Acids Res 10:2851–2864
Yapwattanaphun C, Subhadrabandhu S, Honsho C, Yonemori K (2004) Phylogenetic relationship of mangosteen and several wild relatives revealed by ITS sequence data. J Am Soc Hortic Sci. 129; 368–373. https://doi.org/10.21273/JASHS.129.3.0368
Liu Z, Ni Y, Liu B (2016) Genetic relationships of several Garcinia species (Clusiaceae) revealed by its sequence DATA. IESRJ 2:11–15
Zietkiewicz E, Rafalski A, Labuda D (1994) Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 20:176–183. https://doi.org/10.1006/geno.1994.1151
Charters YM, Robertson A, Wilkinson MJ, Ramsay G (1996) PCR analysis of oilseed rape cultivars (Brassica napus L. ssp. oleifera) using 5′-anchored simple sequence repeat (SSR) primers. Theor Appl Genet 92:442–447. https://doi.org/10.1007/s001220050147
Sikdar B, Bhattacharya M, Mukherjee A, Banerjee A, Ghosh E, Ghosh B, Roy SC (2010) Genetic diversity in important members of Cucurbitaceae using isozyme, RAPD and ISSR markers. Biol Plant 54:135–140. https://doi.org/10.1007/s10535-010-0021-3
Caetano-Anolles G (1994) MAAP: a versatile and universal tool for genome analysis. Plant Mol Biol 25:1011–1026. https://doi.org/10.1007/BF00014674
Parsons TJ, Muniec DS, Sullivan K, Woodyatt N, Alliston-Greiner R, Wilson MR, Holland MM (1997) A high observed substitution rate in the human mitochondrial DNA control region. Nature Genet 15:363–368. https://doi.org/10.1038/ng0497-363
Parthasarathy U, Nandakishore OP, Babu KN, Kumar S, Parthasarathy VA (2013) Comparative effectiveness of inter-simple sequence repeat and randomly amplified polymorphic DNA markers to study genetic diversity of Indian Garcinia. Afr J Biotechnol 12:6443–6451. https://doi.org/10.5897/AJB2013.13053
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed 2:225–238. https://doi.org/10.1007/BF00564200
Anerao JS, Jha V, Korgaonkar L, Devi P, Desai N. (2016) Dissecting genetic diversity in Garcinia xanthochymus using ISSR and RAPD markers. J Plant Breed Genet 4:69–76. http://krishi.icar.gov.in/Publication/handle/123456789/11232
dos Santos AF, Pacheco MV, de Almeida VF, dos Santos FC, Félix FC, das Chagas, KPT, (2016) ISSR molecular markers for the study of the genetic diversity of Mimosa caesalpiniaefolia Bent. IDESIA 34:47–52. https://doi.org/10.4067/S0718-34292016000300007
Acknowledgements
The authors express sincere gratitude to ICMR, New Delhi for financial support (No. 59/14/2011/BMS/TRM, dt.25.03.2015).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Significance Statement
The present study highlighted the phylogenetic analysis of five Garcinia species using ITS1 regions. The study also revealed the genetic similarity and variability among the 42 genotypes of Garcinia species using ISSR molecular marker. It was found that among the five species Garcinia dulcis and Garcinia xanthochymus are genetically similar both in ITS1 sequences comparison and the ISSR marker profiling.
Rights and permissions
About this article
Cite this article
Gogoi, N., Gogoi, A., Neog, B. et al. “Phylogenetic Analysis and Genetic Diversity of Garcinia Species Using ITS Region and ISSR Markers”. Proc. Natl. Acad. Sci., India, Sect. B Biol. Sci. 91, 343–351 (2021). https://doi.org/10.1007/s40011-021-01227-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40011-021-01227-0