Abstract
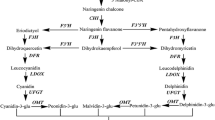
The homologous anthocyanin2 (AN2) genes were identified and structurally characterized in two cultivars of hot pepper Capsicum chinense with different fruit exocarp coloration (purple and yellow). In the analyzed cultivars, the anthocyanin content and the expression pattern of the CcAN2 gene and anthocyanin pathway structural genes DFR and UFGT, regulated by AN2, were determined in the vegetative and reproductive organs, including fruit tissues at different stages of its development. A positive correlation between transcription levels of CcAN2, CcDFR, and CcUFGT was shown. A direct link between transcription levels of the analyzed genes and the anthocyanin content was observed in fruit peel, but not in the leaves. It was assumed that, in the vegetative tissues of the studied cultivars, the AN2, DFR, and UFGT genes can participate in plant photoprotection, and the visible purple color due to the anthocyanin accumulation may be associated with the regulatory activity of other MBW genes.




Similar content being viewed by others
REFERENCES
Gómez-García, M.R. and Ochoa-Alejo, N., Biochemistry and molecular biology of carotenoid biosynthesis in chili peppers (Capsicum spp.), Int. J. Mol. Sci., 2013, vol. 14, no. 9, pp. 19025—19053. https://doi.org/10.3390/ijms140919025
Kong, J.M., Chia, L.S., Goh, N.K., et al., Analysis and biological activities of anthocyanins, Phytochemistry, 2003, vol. 64, pp. 923—933. https://doi.org/10.1016/S0031-9422(03)00438-2
Liu, Y., Tikunov, Y., Schouten, R.E., et al., Anthocyanin biosynthesis and degradation mechanisms in solanaceous vegetables: a review, Front. Chem., 2018, vol. 6, article 52. https://doi.org/10.3389/fchem.2018.00052
Naing, A.H. and Kim, C.K., Roles of R2R3-MYB transcription factors in transcriptional regulation of anthocyanin biosynthesis in horticultural plants, Plant Mol. Biol., 2018, vol. 98, pp. 1—18. https://doi.org/10.1007/s11103-018-0771-4
Albert, N.W., Lewis, D.H., Zhang, H., et al., Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning, Plant J., 2011, vol. 65, pp. 771—784. https://doi.org/10.1111/j.1365-313X.2010.04465.x
Ramsay, N.A. and Glover, B.J., MYB–bHLH–WD40 protein complex and the evolution of cellular diversity, Trends Plant Sci., 2005, vol. 10, no. 2, pp. 63—70.
Koes, R., Verweij, W., and Quattrocchio, F., Flavonoids: a colorful model for the regulation and evolution of biochemical pathways, Trends Plant Sci., 2005, vol. 10, no. 5, pp. 236—242.
Lai, Y., Li, H., and Yamagishi, M., A review of target gene specificity of flavonoid R2R3-MYB transcription factors and a discussion of factors contributing to the target gene selectivity, Front. Biol., 2013, vol. 8, no. 6, pp. 577—598. https://doi.org/10.1007/s11515-013-1281-z
Kim, S., Jones, R., Yoo, K.S., and Pike, L.M., The L locus, one of complementary genes required for anthocyanin production in onions (Allium cepa), encodes anthocyanidin synthase, Theor. Appl. Genet., 2005, vol. 111, pp. 120—127. https://doi.org/10.1007/s00122-005-2000-1
Zong, Y., Zhu, X., Liu, Z., et al., Functional MYB transcription factor encoding gene AN2 is associated with anthocyanin biosynthesis in Lycium ruthenicum Murray, BMC Plant Biol., 2019, vol. 19, article 169.
Kobayashi, S., Yamamoto, N.G., and Hirochika, H., Association of VvmybA1 gene expression with anthocyanin production in grape (Vitis vinifera) skin–color mutants, J. Jpn. Soc. Hortic. Sci., 2005, vol. 74, pp. 196—203.
Strygina, K.V., Kochetov, A.V., and Khlestkina, E.K., Genetic control of anthocyanin pigmentation of potato tissues, BMC Genet., 2019, vol. 20, no. S.1, article 27. https://doi.org/10.1186/s12863-019-0728-x
Quattrocchio, F., Wing, J., van der Woude, K., et al., Molecular analysis of the anthocyanin2 gene of Petunia and its role in the evolution of flower color, Plant Cell, 1999, vol. 11, pp. 1433—1444. https://doi.org/10.1105/tpc.11.8.1433
Kiferle, C., Fantini, E., Bassolino, L., et al., Tomato R2R3-MYB proteins SlANT1 and SlAN2: same protein activity, different roles, PLoS One, 2015, vol. 10, no. 8, article e0136365. https://doi.org/10.1371/journal.pone.0136365
Liu, Y., Lin-Wang, K., Espley, R.V., et al., Functional diversification of the potato R2R3 MYB anthocyanin activators AN1, MYBA1, and MYB113 and their interaction with basic helix-loop-helix cofactors, J. Exp. Bot., 2016, vol. 67, no. 8, pp. 2159—2176. https://doi.org/10.1093/jxb/erw014
Borovsky, Y., Oren-Shamir, M., Ovadia, R., et al., The A locus that controls anthocyanin accumulation in pepper encodes a MYB transcription factor homologous to Anthocyanin2 of Petunia,Theor. Appl. Genet., 2004, vol. 109, pp. 23—29. https://doi.org/10.1007/s00122-004-1625-9
Zhang, Y., Hu, Z., Chu, G., et al., Anthocyanin accumulation and molecular analysis of anthocyanin biosynthesis-associated genes in eggplant (Solanum melongena L.), J. Agric. Food Chem., 2014, vol. 62, no. 13, pp. 2906—2912.
Stommel, J.R. and Dumm, J.M., Coordinated regulation of biosynthetic and regulatory genes coincides with anthocyanin accumulation in developing eggplant fruit, J. Am. Soc. Hortic. Sci., 2015, vol. 140, no. 2, pp. 129—135. https://doi.org/10.21273/JASHS.140.2.129
Moscone, E.A., Scaldaferro, M.A., Grabiele, M., et al., The evolution of chili peppers (Capsicum—Solanaceae): a cytogenetic perspective, Acta Hortic., 2007, vol. 745, pp. 137—170. https://doi.org/10.17660/ActaHortic.2007.745.5
García, C.C., Barfuss, M.H., Sehr, E.M., et al., Phylogenetic relationships, diversification and expansion of chili peppers (Capsicum, Solanaceae), Ann. Bot., 2016, vol. 118, no. 1, pp. 35—51. https://doi.org/10.1093/aob/mcw079
Stommel, J.R., Lightbourn, G.J., Winkel, B.S., and Griesbach, R.J., Transcription factor families regulate the anthocyanin biosynthetic pathway in Capsicum annuum,J. Am. Soc. Hortic. Sci., 2009, vol. 134, pp. 244—251.
Aza-González, C., Herrera-Isidrón, L., Núñez-Palenius, H.G., et al., Anthocyanin accumulation and expression analysis of biosynthesis-related genes during chili pepper fruit development, Biol. Plant., 2013, vol. 57, pp. 49—55. https://doi.org/10.1007/s10535-012-0265-1
Zhang, Z., Li, D.W., Jin, J.H., et al., VIGS approach reveals the modulation of anthocyanin biosynthetic genes by CaMYB in chili pepper leaves, Front Plant Sci., 2015, vol. 6, article 500. https://doi.org/10.3389/fpls.2015.00500
Solovchenko, A.E., Chivkunova, O.B., Merzlyak, M.N., and Reshetnikova, I.V., A spectrophotometric analysis of pigments in apples, Russ. J. Plant Physiol., 2001, vol. 48, no. 5, pp. 693—700.
Zimmermann, I.M., Heim, M.A., Weisshaar, B., and Uhrig, J.F., Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins, Plant J., 2004, vol. 40, pp. 22—34.
Bemer, M., Karlova, R., Ballester, A.R., et al., The tomato FRUITFULL homologs TDR4/FUL1 and MBP7/FUL2 regulate ethylene-independent aspects of fruit ripening, Plant Cell, 2012, vol. 24, no. 11, pp. 4437—4451. https://doi.org/10.1105/tpc.112.103283
Lin-Wang, K., Bolitho, K., Grafton, K., et al., An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae, BMC Plant Biol., 2010, vol. 10, article 50. https://doi.org/10.1186/1471-2229-10-50
Emiliani, G., Fondi, M., Fani, R., and Gribaldo, S., A horizontal gene transfer at the origin of phenylpropanoid metabolism: a key adaptation of plants to land, Biol. Direct, 2009, vol. 4, article 7. https://doi.org/10.1186/1745-6150-4-7
Takeda, J., Nakata, R., Ueno, H., et al., Possible involvement of a tetrahydrobiopterin in photoreception for UV-B-induced anthocyanin synthesis in carrot, Photochem. Photobiol., 2014, vol. 90, no. 5, pp. 1043—1049.
Grotewold, E., The genetics and biochemistry of floral pigments, Annu. Rev. Plant Biol., 2006, vol. 57, pp. 761—780.
Särkinen, T., Bohs, L., Olmstead, R.G., and Knapp, S., A phylogenetic framework for evolutionary study of the nightshades (Solanaceae): a dated 1000-tip tree, BMC Evol. Biol., 2013, vol. 13, article 214. https://doi.org/10.1186/1471-2148-13-214
Funding
This work was supported by the Russian Science Foundation, project no. 19-16-00016, and partially by the Ministry of Science and Higher Education of the Russian Federation (M.A. Filyushin, phylogenetic analysis of the AN2 sequences of Solanaceae), and was performed using the experimental climate control facility in the Institute of Bioengineering, Research Centre of Biotechnology, Russian Academy of Sciences.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare no conflict of interest.
This article contains no studies with use of people or animals as objects.
Additional information
Translated by M. Bibov
Rights and permissions
About this article
Cite this article
Filyushin, M.A., Dzhos, E.A., Shchennikova, A.V. et al. Expression Features of the Transcription Factor Gene anthocyanin2 and Its Effect on the Anthocyanin Content in Capsicum chinense Jacq. Cultivars with Different Fruit Coloration. Russ J Genet 56, 1203–1211 (2020). https://doi.org/10.1134/S1022795420090069
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795420090069