Abstract
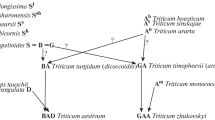
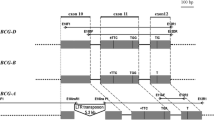
Polyploidy is the major mechanism of speciation in flowering plants. All genomes of ancient species that are the progenitors of extant plant species experienced polyploidization. Three consecutive stages of polyploidization, i.e., ancient polyploidization, tetra-, and hexaploidization, resulted in the emergence of modern allohexaploid bread wheat Triticum aestivum L. with the BBAADD genome. Polyploidization and subsequent stabilization of the polyploid genome of T. aestivum led, on one hand, to cytological diploidization and, on the other hand, to structural and functional asymmetry of its three subgenomes. In recent years, there has been a sharp increase in the data accumulation on the origin and structure of the bread wheat genomes a result of analysis of genomes and transcripomes of natural and synthetic wheats using modern mapping and sequencing methods. This review provides up-to-date information on the peculiarities of the T. aestivum genome reorganization, which affected its structure and functioning.
Similar content being viewed by others
References
Chen, Z.J., Molecular mechanisms of polyploidy and hybrid vigor, Trends Plant Sci., 2010, vol. 15, pp. 57–71. doi 10.1016/j.tplants.2009.12.003
Mayfield, D., Chen, Z.J., and Pires, J.C., Epigenetic regulation of flowering time in polyploids, Curr. Opin. Plant Biol., 2011, vol. 14, pp. 174–178. doi 10.1016/j.pbi.2011.03.008
Soltis, D.E., Albert, V.A., Leebens-Mack, J., et al., Polyploidy and angiosperm diversification, Am. J. Bot., 2009, vol. 96, no. 1, pp. 336–348. doi 10.3732/ajb.0800079
Stebbins, G.L., Types of polyploids: their classification and significance, Adv. Genet., 1947, vol. 1, pp. 403–429.
Madlung, A. and Wendel, J.F., Genetic and epigenetic aspects of polyploid evolution in plants, Cytogenet. Genome Res., 2013, vol. 140, nos. 2–4, pp. 270–285. doi 10.1159/000351430
Otto, S.P. and Whitton, J., Polyploid incidence and evolution, Annu. Rev. Genet., 2000, vol. 34, pp. 401–437. doi 10.1146/annurev.genet.34.1.401
Cui, L., Wall, P.K., Leebens-Mack, J.H., et al., Widespread duplications throughout the history of flowering plants, Genome Res., 2006, vol. 16, pp. 738–749. doi 10.1101/gr.4825606
Leitch, A.R. and Leitch, I.J., Genomic plasticity and the diversity of polyploid plants, Science, 2008, vol. 320, no. 5875, pp. 481–483. doi 10.1126/science. 1153585
Amborella Genome Project, The Amborella genome and the evolution of flowering plants, Science, 2013, vol. 342, p. 1241089. doi 10.1126/science.1241089
Renny-Byfield, S. and Wendel, J.F., Doubling down on genomes: polyploidy and crop plants, Am. J. Bot., 2014, vol. 101, no. 10, pp. 1711–1725. doi 10.3732/ajb.1400119
Garsmeur, O., Schnable, J.C., Almeida, A., et al., Two evolutionarily distinct classes of paleopolyploidy, Mol. Biol. Evol., 2013, vol. 31, pp. 448–454. doi 10.1093/molbev/mst230
Kim, E.S., Bol’sheva, N.L., Samatadze, T.E., et al., The unique genome of two-chromosome grasses Zingeria and Colpodium, its origin, and evolution, Russ. J. Genet., 2009, vol. 45, no. 11, pp. 1329–1337. doi 10.1134/S1022795409110076
Rodionov, A.V., Polyploidy and interspecific hybridization in the evolution of flowering plants, Vavilovskii Zh. Genet. Sel., 2013, vol. 17, no. 4/2, pp. 916–929.
Salse, J., Bolot, S., Throude, M., et al., Identification and characterization of conserved duplications between rice and wheat provide new insight into grass genome evolution, Plant Cell, 2008, vol. 20, pp. 11–24. doi 10.1105/tpc.107.056309
Murat, F., Xu, J.H., Tannier, E., et al., Ancestral grass karyotype reconstruction unravels new mechanisms of genome shuffling as a source of plant evolution, Genome Res., 2010, vol. 20, no. 11, pp. 1545–1557. doi 10.1101/gr.109744.110
Murat, F., Zhang, R., Guizard, S., et al., Shared subgenome dominance following polyploidization explains grass genome evolutionary plasticity from a seven protochromosome ancestor with 16K protogenes, Genome Biol. Evol., 2014, vol. 6, pp. 12–33. doi 10.1093/gbe/evt200
Pont, C., Murat, F., Guizard, S., et al., Wheat syntenome unveils new evidences of contrasted evolutionary plasticity between paleo-and neoduplicated subgenomes, Plant J., 2013, vol. 76, no. 6, pp. 1030–1044. doi 10.1111/tpj.12366
Shcherban’, A.B., The reorganization of plant genomes during allopolyploidization, Russ. J. Genet.: Appl. Res., 2013, vol. 3, no. 6, pp. 444–450. https://doi.org/10.1134/S2079059713060087.
Badaeva, E.D. and Salina, E.A., Genome structure and the chromosome analysis of plants, Vavilovskii Zh. Genet. Sel., 2013, vol. 17, no. 4/2, pp. 1017–1043.
International Wheat Genome Sequencing Consortium, A chromosome based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome, Science, 2014, vol. 345, no. 80, pp. 1250092–1250092. doi 10.1126/science.1251788
Feldman, M., Levy, A.A., Fahima, T., and Korol, A., Genomic asymmetry in allopolyploid plants: wheat as a model, J. Exp. Bot., 2012, vol. 63, no. 14, pp. 5045–5059. doi 10.1093/jxb/ers192
Leach, L.J., Belfield, E.J., Jiang, C., et al., Patterns of homoeologous gene expression shown by RNA sequencing in hexaploid bread wheat, BMC Genomics, 2014, vol. 15, p. 276. doi 10.1186/1471-2164-15-276
Matsuoka, Y., Evolution of polyploid Triticum wheats under cultivation: the role of domestication, natural hybridization, and allopolyploid speciation in their diversification, Plant Cell Physiol., 2011, vol. 52, pp. 750–764. doi 10.1093/pcp/pcr018
Salina, E.A., Lim, K.Y., Badaeva, E.D., et al., Phylogenetic reconstruction of Aegilops section Sitopsis and the evolution of tandem repeats in the diploids and derived wheat polyploids, Genome, 2006, vol. 49, no. 8, pp. 1023–1035. doi 10.1139/g06-050
Salse, J., Chagué, V., Bolot, S., et al., New insights into the origin of the B genome of hexaploid wheat: evolutionary relationships at the SPA genomic region with the S genome of the diploid relative Aegilops speltoides, BMC Genomics, 2008, vol. 9, p. 555. doi 1471-2164/9/555
Marcussen, T., Sandve, S.R., Heier, L., et al., Ancient hybridizations among the ancestral genomes of bread wheat, Science, 2014, vol. 45, no. 6419, p. 1250092. doi 10.1126/science.1250092
Jones, N. and Pasakinskien, I., Genome conflict in the Gramineae, New Phytol., 2005, vol. 165, pp. 391–410. doi 10.1111/j.1469-8137.2004.01225.x
Flavell, R.B., Rimpau, J., and Smith, D.B., Repeated sequence DNA relationships in 4 cereal genomes, Chromosoma, 1977, vol. 63, no. 3, pp. 205–222.
Wanjugi, H., Coleman-Derr, D., Huo, N., et al., Rapid development of PCR-based genome-specific repetitive DNA junction markers in wheat, Genome, 2009, vol. 52, no. 6, pp. 576–587. doi 10.1139/g09-033
Sears, E.R., Wheat cytogenetics, Annu. Rev. Genet., 1969, vol. 3, pp. 451–468.
Levy, A.A. and Feldman, M., Genetic and epigenetic reprogramming of the wheat genome upon allopolyploidization, Biol. J. Linn. Soc., 2004, vol. 82, no. 4, pp. 607–613. doi 10.1111/j.1095-8312.2004.00346.x
Wicker, T., Mayer, K.F., Gundlach, H., et al., Frequent gene movement and pseudogene evolution is common to the large and complex genomes of wheat, barley, and their relatives, Plant Cell, 2011, vol. 23, no. 5, pp. 1706–1718. doi 10.1105/tpc.111.086629
Feldman, M., Liu, B., Segal, G., et al., Rapid elimination of low-copy DNA sequences in polyploid wheat: a possible mechanism for differentiation of homoeologous chromosomes, Genetics, 1997, vol. 147, pp. 1381–1387.
Ozkan, H., Levy, A.A., and Feldman, M., Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops–Triticum) group, Plant Cell, 2001, vol. 13, pp. 1735–1747. doi 10.1105/TPC.010082
Ozkan, H., Tuna, M., and Arumuganathan, K., Nonadditive changes in genome size during allopolyploidization in the wheat (Aegilops–Triticum) group, J. Hered., 2003, vol. 94, pp. 260–264. doi 10.1093/jhered/esg053
Han, F.P., Fedak, G., Quellet, T., and Liu, B., Rapid genomic exchanges in interspecific and intergeneric hybrids and allopolyploids of Triticeae, Genome, 2003, vol. 46, pp. 716–723. doi 10.1139/g03-049
Salina, E.A., Numerova, O.M., Ozkan, H., and Feldman, M., Alterations in subtelomeric tandem repeats during early stages of allopolyploidy in wheat, Genome, 2004, vol. 47, pp. 860–867. doi 10.1139/g04-044
Eilam, T., Anikster, Y., Millet, E., et al., Nuclear DNA amount and genome downsizing in natural and synthetic allopolyploids of the genera Aegilops and Triticum, Genome, 2008, vol. 51, no. 8, pp. 616–627. doi 10.1139/G08-043
Feldman, M. and Levy, A.A., Genome evolution in allopolyploid wheat–a revolutionary reprogramming followed by gradual changes, J. Genet. Genomics, 2009, vol. 36, pp. 511–518. doi 10.1016/S1673-8527(08)60142-3
Raats, D., Frenkel, Z., Krugman, T., et al., The physical map of wheat chromosome 1BS provides insights into its gene space organization and evolution, Genome Biol., 2013, vol. 14, p. R138. doi 10.1186/gb-2013-14-12-r138
Philippe, R., Paux, E., Bertin, I., et al., A high density physical map of chromosome 1BL supports evolutionary studies, map-based cloning and sequencing in wheat, Genome Biol., 2013, vol. 14, p. R64. doi 10.1186/gb-2013-14-6-r64
Sehgal, S.K., Li, W., Rabinowicz, P.D., et al., Chromosome arm-specific BAC end sequences permit comparative analysis of homoeologous chromosomes and genomes of polyploid wheat, BMC Plant Biol., 2012, vol. 12, p. 64. doi 10.1186/1471-2229-12-64
Choulet, F., Wicker, T., Rustenholz, C., et al., Megabase level sequencing reveals contrasted organization and evolution patterns of the wheat gene and transposable element spaces, Plant Cell, 2010, vol. 22, no. 6, pp. 1686–1701. doi 10.1105/tpc.110.074187
Choulet, F., Alberti, A., Theil, S., et al., Structural and functional partitioning of bread wheat chromosome 3B, Science, 2014, vol. 345, no. 6194, p. 1249721. doi 10.1126/science.1249721
Pingault, L., Choulet, F., Alberti, A., et al., Deep transcriptome sequencing provides new insights into the structural and functional organization of the wheat genome, Genome Biol., 2015, vol. 16, p. 29. doi 10.1186/s13059-015-0601-9
Bartoš, J., Vlček, C., Choulet, F., et al., Intraspecific sequence comparisons reveal similar rates of non-collinear gene insertion in the B and D genomes of bread wheat, BMC Plant Biol., 2012, vol. 12, p. 155. doi 10.1186/1471-2229-12-155
Hernandez, P., Martis, M., Dorado, G., et al., Nextgeneration sequencing and syntenic integration of flow-sorted arms of wheat chromosome 4A exposes the chromosome structure and gene content, Plant J., 2012, vol. 69, no. 3, pp. 377–386. doi 10.1111/j.1365-313X.2011.04808.x
Helguera, M., Rivarola, M., Clavijo, B., et al., New insights into the wheat chromosome 4D structure and virtual gene order, revealed by survey pyrosequencing, Plant Sci., 2015, vol. 233, pp. 200–212. doi 10.1016/j.plantsci.2014.12.004
Vitulo, N., Albiero, A., Forcato, C., et al., First survey of the wheat chromosome 5A composition through a next generation sequencing approach, PLoS One, 2011, vol. 6, no. 10. e26421. doi 10.1371/journal. pone.0026421
Sergeeva, E.M., Afonnikov, D.A., Koltunova, M.K., et al., Common wheat chromosome 5B composition analysis using low-coverage 454 sequencing, Plant Genome, 2014, vol. 7, pp. 1–16. doi 10.3835/plantgenome2013.10.0031
Lucas, S.J., Akpınar, B.A., Šimková, H., et al., Nextgeneration sequencing of flow-sorted wheat chromosome 5D reveals lineage-specific translocations and widespread gene duplications, BMC Genomics, 2014, vol. 15, p. 1080. doi 10.1186/1471-2164-15-1080
Tanaka, T., Kobayashi, F., Joshi, G.P., et al., Nextgeneration survey sequencing and the molecular organization of wheat chromosome 6B, DNA Res., 2014, vol. 1, no. 2, pp. 103–114. doi 10.1093/dnares/dst041
Kobayashi, F., Wu, J., Kanamori, H., et al., A highresolution physical map integrating an anchored chromosome with the BAC physical maps of wheat chromosome 6B, BMC Genomics, 2015, vol. 16, p. 595. doi 10.1186/s12864-015-1803-y
Berkman, P.J., Skarshewski, A., Lorenc, M.T., et al., Sequencing and assembly of low copy and genic regions of isolated Triticum aestivum chromosome arm 7DS, Plant Biotechnol. J., 2011, vol. 9, no. 7, pp. 768–775. doi 10.1111/j.1467-7652.2010.00587.x
Berkman, P.J., Skarshewski, A., Manoli, S., et al., Sequencing wheat chromosome arm 7BS delimits the 7BS/4AL translocation and reveals homoeologous gene conservation, Theor. Appl. Genet., 2012, vol. 124, no. 3, pp. 423–432. doi 10.1007/s00122-011-1717-2
Breen, J., Wicker, T., Shatalina, M., et al., A physical map of the short arm of wheat chromosome 1A, PLoS One, 2013, vol. 8, no. 11. e80272. doi 10.1371/journal. pone.0080272
Akpinar, B.A., Magni, F., Yuce, M., et al., The physical map of wheat chromosome 5DS revealed gene duplications and small rearrangements, BMC Genomics, 2015, vol. 16, p. 453. doi 10.1186/s12864-015-1641-y
Conley, E.J., Nduati, V., Gonzalez-Hernandez, J.L., et al., A 2600-locus chromosome bin map of wheat homoeologous group 2 reveals interstitial gene-rich islands and collinearity with rice, Genetics, 2004, vol. 168, no. 2, pp. 625–637. doi 10.1534/genetics. 104.034801
Balcárková, B., Frenkel, Z., Škopová, M., et al., A high resolution radiation hybrid map of wheat chromosome 4A, Front. Plant Sci., 2017, vol. 7, p. 2063. doi 10.3389/fpls.2016.02063
Tiwari, V.K., Heesacker, A., Riera-Lizarazu, O., et al., A whole-genome radiation hybrid mapping resource of hexaploid wheat, Plant J., 2016, vol. 86, pp. 195–207. doi 10.1111/tpj.13153
Brenchley, R., Spannagl, M., Pfeifer, M., et al., Analysis of the bread wheat genome using whole-genome shotgun sequencing, Nature, 2012, vol. 491, no. 7426, pp. 705–710. doi 10.1038/nature11650
Bolot, S., Abrouk, M., Masood-Quraishi, U., et al., The ‘inner circle’ of the cereal genomes, Curr. Opin. Plant Biol., 2009, vol. 12, pp. 119–125. doi 10.1016/j.pbi.2008.10.011
Mayer, K.F., Martis, M., Hedley, P.E., et al., Unlocking the barley genome by chromosomal and comparative genomics, Plant Cell, 2011, vol. 23, pp. 1249–1263. doi 10.1105/tpc.110.082537
Martis, M.M., Zhou, R., Haseneyer, G., et al., Reticulate evolution of the rye genome, Plant Cell, 2013, vol. 25, pp. 3685–3698. doi 10.1105/tpc.113.114553
Naranjo, T., Roca, A., Goicoechea, P.G., and Giraldez, R., Arm homoeology of wheat and rye chromosomes, Genome, 1987, vol. 29, no. 6, pp. 873–882. doi 10.1139/g87-149
Devos, K.M., Dubcovsky, J., Dvorak, J., et al., Structural evolution of wheat chromosomes 4a, 5a, and 7b and its impact on recombination, Theor. Appl. Genet., 1995, vol. 91, no. 2, pp. 282–288. doi 10.1007/BF00220890
Li, W., Challa, G.S., Zhu, H., and Wei, W., Recurrence of chromosome rearrangements and reuse of DNA breakpoints in the evolution of the Triticeae genomes, G3 (Bethesda), 2016, vol. 6, no. 12, pp. 3837–3847. doi 10.1534/g3.116.035089
Qi, L.L., Chen, P.D., Liu, D.J., and Gill, B.S., Homoeologous relationships of Haynaldia villosa chromosomes with those of Triticum aestivum as revealed by RFLP analysis, Genes Genet. Syst., 1999, vol. 74, pp. 77–82. doi 10.1266/ggs.74.77
Akhunov, E.D., Goodyear, A.W., Geng, S., et al., The organization and rate of evolution of wheat genomes are correlated with recombination rates along chromosome arms, Genome Res., 2003, vol. 13, no. 5, pp. 753–763. doi 10.1101/gr.808603GR-8086R
Badaeva, E.D., Dedkova, O.S., Gay, G., et al., Chromosomal rearrangements in wheat: their types and distribution, Genome, 2007, vol. 50, no. 10, pp. 907–926. doi 10.1139/g07-072
Zhang, H., Bian, Y., Gou, X., et al., Persistent wholechromosome aneuploidy is generally associated with nascent allohexaploid wheat, Proc. Natl. Acad. Sci. U.S.A., 2013, vol. 110, pp. 3447–3452. doi 10.1073/pnas.1300153110
Roder, M.S., Korzun, V., Wendehake, K., et al., A microsatellite map of wheat, Genetics, 1998, vol. 149, pp. 2007–2023.
Akhunov, E.D., Akhunova, A.R., Anderson, O.D., et al., Nucleotide diversity maps reveal variation in diversity among wheat genomes and chromosomes, BMC Genomics, 2010, vol. 11, p. 702. doi 10.1186/1471-2164-11-702
Allen, A.M., Barker, G.L., Berry, S.T., et al., Transcript-specific, single-nucleotide polymorphism discovery and linkage analysis in hexaploid bread wheat (Triticum aestivum L.), Plant Biotechnol. J., 2011, vol. 9, pp. 1086–1099. doi 10.1111/j.1467-7652.2011. 00628.x
Berkman, P.J., Visendi, P., Lee, H.C., et al., Dispersion and domestication shaped the genome of bread wheat, Plant Biotechnol. J., 2013, vol. 11, no. 5, pp. 564–571. doi 10.1111/pbi.12044
Bottley, A. and Koebner, R.M.D., Variation for homoeologous gene silencing in hexaploid wheat, Plant J., 2008, vol. 56, pp. 297–302. doi 10.1186/1471-2199-8-65
Ge, X.-H., Ding, L., and Li, Z.-Y., Nucleolar dominance and different genome behaviors in hybrids and allopolyploids, Plant Cell Rep., 2013, vol. 32, pp. 1661–1673. doi 10.1007/s00299-013-1475-5
Pfeifer, M., Kugler, K.G., Sandve, S.R., et al., Genome interplay in the grain transcriptome of hexaploid bread wheat, Science, 2014, vol. 345, no. 6194, p. 1250091. doi 10.1126/science.1250091
Pumphrey, M., Bai, J., Laudencia-Chingcuanco, D., et al., Nonadditive expression of homoeologous genes is established upon polyploidization in hexaploid wheat, Genetics, 2009, vol. 181, pp. 1147–1157. doi 10.1534/genetics.108.096941
Sears, E.R., An induced mutant with homoeologous pairing in common wheat, Can. J. Genet. Cytol., 1977, vol. 19, pp. 585–593.
Moore, G. and Shaw, P., Improving the chances of finding the right partner, Curr. Opin. Genet. Dev., 2009, vol. 19, no. 2, pp. 99–104. doi 10.1016/j.gde.2009.02.006
Luo, M.C., Dubcovsky, J., and Dvorák, J., Recognition of homeology by the wheat Ph1 locus, Genetics, 1996, vol. 144, no. 3, pp. 1195–1203.
Giorgi, B., A homoeologous pairing mutant isolated in Triticum durum cv. Cappelli, Mutat. Breed. Newslett., 1978, vol. 11, pp. 4–5.
Sears, E.R., Genetic control of chromosome pairing in common wheat, Annu. Rev. Genet., 1976, vol. 10, pp. 31–51.
Feldman, M., The effect of chromosomes 5B, 5D, and 5A on chromosomal pairing in Triticum aestivum, Proc. Natl. Acad. Sci. U.S.A., 1966, vol. 55, no. 1, pp. 447–453.
Riley, R., Chapman, V., Young, R.M., and Belfield, A.M., Control of meiotic chromosome pairing by the chromosomes of homoeologous group 5 of Triticum aestivum, Nature, 1966, vol. 212, pp. 1475–1477. doi 10.1038/2121475a0
Boden, S.A., Shadiac, N., Tucker, E.J., et al., Expression and functional analysis of TaASY1 during meiosis of bread wheat (Triticum aestivum), BMC Mol. Biol., 2007, vol. 8, p. 65. doi 10.1186/1471-2199-8-65
Boden, S.A., Langridge, P., Spangenberg, G., and Able, J.A., TaASY1 promotes homologous chromosome interactions and is affected by deletion of Ph1, Plant J., 2009, vol. 57, no. 3, pp. 487–497. doi 10.1111/j.1365-313X.2008.03701.x
Griffiths, S., Sharp, R., Foote, T.N., et al., Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat, Nature, 2006, vol. 439, no. 7077, pp. 749–752. doi 10.1038/nature04434
Yousafzai, F.K., Al-Kaff, N., and Moore, G., Structural and functional relationship between the Ph1 locus protein 5B2 in wheat and CDK2 in mammals, Funct. Integr. Genomics, 2010, vol. 10, pp. 157–166. doi 10.1007/s10142-010-0170-7
Al-Kaff, N., Knight, E., Bertin, I., et al., Detailed dissection of the chromosomal region containing the Ph1locus in wheat Triticum aestivum: with deletion mutants and expression profiling, Ann. Bot., 2008, vol. 101, pp. 863–872. doi 10.1093/aob/mcm252
Greer, E., Martín, A.C., Pendle, A., et al., The Ph1 locus suppresses Cdk2-type activity during premeiosis and meiosis in wheat, Plant Cell, 2012, vol. 24, no. 1, pp. 152–162. doi 10.1105/tpc.111.094771
Sidhu, G.K., Rustgi, S., Shafqat, M.N., et al., Fine structure mapping of a gene-rich region of wheat carrying Ph1, a suppressor of crossing over between homoeologous chromosomes, Proc. Natl. Acad. Sci. U.S.A., 2008, vol. 105, no. 15, pp. 5815–5820. doi 10.1073/pnas.0800931105
Bhullar, R., Nagarajan, R., Bennypaul, H., et al., Silencing of a metaphase I-specific gene results in a phenotype similar to that of the Pairing homeologous 1 (Ph1) gene mutations, Proc. Natl. Acad. Sci. U.S.A., 2014, vol. 111, no. 39, pp. 14187–14192. doi 10.1073/pnas.1416241111
Glover, N.M., Daron, J., Pingault, L., et al., Smallscale gene duplications played a major role in the recent evolution of wheat chromosome 3B, Genome Biol., 2015, vol. 16, p. 188. doi 10.1186/s13059-015-0754-6
Salina, E.A., Sergeeva, E.M., Adonina, I.G., et al., The impact of Ty3-gypsy group LTR retrotransposons Fatima on B-genome specificity of polyploid wheats, BMC Plant Biol., 2011, vol. 11, p. 99. doi 10.1186/1471-2229-11-99
Hegarty, M.J. and Hiscock, S.J., Genomic clues to the evolutionary success of review polyploid plants, Curr. Biol., 2008, vol. 18, no. 10, pp. R435–R444. doi 10.1016/j.cub.2008.03.043
Yang, C., Zhao, L., Zhang, H., et al., Evolution of physiological responses to salt stress in hexaploid wheat, Proc. Natl. Acad. Sci. U.S.A., 2014, vol. 111, pp. 11882–11887. doi 10.1073/pnas.1412839111
Han, Y., Xin, M., Huang, K., et al., Altered expression of TaRSL4 gene by genome interplay shapes root hair length in allopolyploid wheat, New Phytol., 2016, vol. 209, no. 2, pp. 721–732. doi 10.1111/nph.13615
Pont, C., Murat, F., Confolent, C., et al., RNA-seq in grain unveils fate of neo-and paleopolyploidization events in bread wheat (Triticum aestivum L.), Genome Biol., 2011, vol. 12, no. 12, p. R119. doi 10.1186/gb-2011-12-12-r119
Wang, Y.P., Cheng, X., Shan, Q.W., et al., Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew, Nat. Biotechnol., 2014, vol. 32, pp. 947–951. doi 10.1038/nbt.2969
Kenan-Eichler, M., Leshkowitz, D., Tal, L., et al., Wheat hybridization and polyploidization results in deregulation of small RNAs, Genetics, 2011, vol. 188, no. 2, pp. 263–272. doi 10.1534/genetics.111.128348
Zhao, N., Zhu, B., Li, M., et al., Extensive and heritable epigenetic remodeling and genetic stability accompany allohexaploidization of wheat, Genetics, 2011, vol. 188, no. 3, pp. 499–510. doi 10.1534/genetics. 111.127688
Zhang, H., Bian, Y., Gou, X., et al., Intrinsic karyotype stability and gene copy number variations may have laid the foundation for tetraploid wheat formation, Proc. Natl. Acad. Sci. U.S.A., 2013, vol. 110, pp. 19466–19471. doi 10.1073/pnas.1319598110
Kashkush, K., Feldman, M., and Levy, A.A., Gene loss, silencing and activation in a newly synthesized wheat allotetraploid, Genetics, 2002, vol. 160, pp. 1651–1659.
Shaked, H., Kashkush, K., Ozkan, H., et al., Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat, Plant Cell, 2001, vol. 13, pp. 1749–1759. doi 10.1105/TPC.010083
Liu, C., Yang, X., Zhang, H., et al., 4, Plant Mol. Biol., 2015, vol. 88, nos. 1–2, pp. 53–64. doi 10.1007/s11103-015-0307-0
Han, F., Fedak, G., Guo, W., and Liu, B., Rapid and repeatable elimination of a parental genome-specific DNA repeat (pGc1R-1a) in newly synthesized wheat allopolyploids, Genetics, 2005, vol. 170, pp. 1239–1245. doi 10.1534/genetics.104.039263
Bento, M., Gustafson, J.P., Viegas, W., and Silva, M., Size matters in Triticeae polyploids: larger genomes have higher remodeling, Genome, 2011, vol. 54, pp. 175–183. doi 10.1139/G10-107
Qi, B., Huang, W., Zhu, B., et al., Global transgenerational gene expression dynamics in two newly synthesized allohexaploid wheat (Triticum aestivum) lines, BMC Biol., 2012, vol. 10, p. 3. doi 10.1186/1741-7007-10-3
Guo, X. and Han, F.P., Asymmetric epigenetic modification and elimination of rDNA sequences by polyploidization in wheat, Plant Cell, 2014, vol. 26, pp. 4311–4327. doi 10.1105/tpc.114.129841
Cantu, D., Vanzetti, L.S., Sumner, A., et al., Small RNAs, DNA methylation and transposable elements in wheat, BMC Genomics, 2010, vol. 11, p. 408. doi 10.1186/1471-2164-11-408
Kraitshtein, Z., Yaakov, B., Khasdan, V., and Kashkush, K., Genetic and epigenetic dynamics of a retrotransposon after allopolyploidization of wheat, Genetics, 2010, vol. 186, pp. 801–812. doi 10.1534/genetics. 110.120790
Ben-David, S., Yaakov, B., and Kashkush, K., Genome-wide analysis of short interspersed nuclear elements SINES revealed high sequence conservation, gene association and retrotranspositional activity in wheat, Plant J., 2013, vol. 76, pp. 201–210. doi 10.1111/tpj.12285
Zhang, H., Zhu, B., Qi, B., et al., Evolution of the BBAA component of bread wheat during its history at the allohexaploid level, Plant Cell, 2014, vol. 26, no. 7, pp. 2761–2776. doi 10.1105/tpc.114.128439
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © D.B. Loginova, O.G. Silkova, 2018, published in Genetika, 2018, Vol. 54, No. 4.
Rights and permissions
About this article
Cite this article
Loginova, D.B., Silkova, O.G. The Genome of Bread Wheat Triticum aestivum L.: Unique Structural and Functional Properties. Russ J Genet 54, 403–414 (2018). https://doi.org/10.1134/S1022795418040105
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795418040105