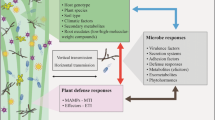
Abstract
In the present work, the potential for the enhancement of the adaptive capacity of microbe–plant systems (MPSs) through the integration of the symbiosis partners’ genomes is considered on the example of different types of symbiotic relationships. The accumulated data on the genetic control of interactions for both the plant and microbe, which are discussed in the paper with respect to signaling genes, suggest that it is the complementarity of genetic determinants that underlies the successful formation of MPSs. A eukaryotic genome with limited information content, which is stable throughout a generation, is complemented by a virtually unlimited prokaryotic metagenome. The microsymbiont’s ability to adapt to different living conditions is based on the restructuring of the accessory genome by different mechanisms, which are likely to be activated under the influence of plants, although the details of such a regulation remain unknown. Features of the genetic control of the interaction, particularly its universal character for different symbionts, allow us to formulate a principle of genome complementarity with respect to interacting organisms and consider it an important factor, an adaptation that enhances the abilities of MPSs for their sustainable development in natural ecosystems and for high plant productivity in agrocenoses.
Similar content being viewed by others
References
FAOSTAT, 2014. http://faostat.fao.org/
Coghlan, A., Eichler, E.E., Oliver, S.G., et al., Chromosome evolution in eukaryotes: a multi-kingdom perspective, Trends Genet., 2005, vol. 21, pp. 673–682.
Hugenholtz, Ph., Exploring prokaryotic diversity in the genomic era, Genome Biol., 2002, vol. 2, no. 3, pp. 0003.1–0003.8.
Daniel, R., The metagenomics of soil, Nat. Rev. Microbiol., 2005, vol. 6, no. 3, pp. 470–478.
Torsvik, V., Goksøyr, J., and Daae, F.L., High diversity in DNA of soil bacteria, Appl. Environ. Microbiol., 1990, vol. 3, no. 56, pp. 782–787.
Morgan, N., The complexity chronicles, Nature, 2014, vol. 510, no. 7505, pp. 338–339.
Margulis, L., Symbiosis in Cell Evolution: Life and Its Environment on the Early Earth, San Francisco: Freeman, 1981.
Tikhonovich, I.A. and Provorov, N.A., From plantmicrobe interactions to symbiogenetics: a universal paradigm for the interspecies genetic integration, Ann. Appl. Biol., 2009, vol. 154, no. 3, pp. 341–350.
Roger, E., Soil science comes to life, Nature, 2013, vol. 501, no. 7468, pp. 18–19.
Belimov, A.A., Dodd, I.C., Hontzeas, N., et al., Rhizosphere bacteria containing 1-aminocyclopropane-1carboxylate deaminase increase yield of plants grown in drying soil via both local and systemic hormone signaling, New Phytol., 2009, vol. 181, no. 2, pp. 413–423.
Berendsen, R.L., Pieterse, C.M., and Bakker, P.A., The rhizosphere microbiome and plant health, Trends Plant Sci., 2012, vol. 17, no. 8, pp. 478–486.
Lundberg, D.S., Lebeis, S.L., Paredes, S.H., et al., Defining the core Arabidopsis thaliana root microbiome, Nature, 2012, vol. 488, no. 7409, pp. 86–90.
Turnbaugh, P.J., Hamady, M., Yatsunenko, T., et al., A core gut microbiome in obese and lean twins, Nature, 2009, vol. 457, no. 7228, pp. 480–484.
Borisov, A.Y., Danilova, T.N., Koroleva, T.A., et al., Pea (Pisum sativum L.) regulatory genes controlling development of nitrogen-fixing nodule and arbuscular mycorrhiza: fundamentals and application, Biologia, 2004, vol. 59, suppl. 13, pp. 137–144.
Borisov, A.Yu., Vasil’chikov, A.G., Voroshilova, V.A., et al., Regulatory genes of garden pea (Pisum sativum L.) controlling the development of nitrogen-fixing nodules and arbuscular mycorrhiza: a review of basic and applied aspects, Appl. Biochem. Microbiol., 2007, vol. 43, no. 3, pp. 237–243.
Borisov, A.Yu., Shtark, O.Yu., Zhukov, V.A., et al., Interaction of legumes with beneficial soil microorganisms: from plant genes to varieties, S-kh. Biol., 2011, no. 3, pp. 41–47.
Tikhonovich, I.A. and Provorov, N.A., Symbiogenetics of plant–microbe interactions, Ekol. Genet., 2003, no. 1, pp. 36–46.
Tikhonovich, I.A. and Provorov, N.A., Agricultural microbiology as the basis of ecologically sustainable agriculture: fundamental and applied aspects, S-kh. Biol., 2011, no. 3, pp. 3–9.
Zhukov, V.A., Shtark, O.Yu., Borisov, A.Yu., and Tikhonovich, I.A., Molecular genetic mechanisms used by legumes to control early stages of mutually beneficial (mutualistic) symbiosis, Russ. J. Genet., 2009, vol. 45, no. 11, pp. 1279–1288.
Zhukov, V.A., Zhernakov, A.I., Ershov, N.I., et al., Regulation of morphogenesis of symbiotic nodules of pea (Pisum sativum L.), identified through transcriptome sequencing, in Tezisy dokladov VIs”ezda Vavilovskogo obshchestva genetikov i selektsionerov (VOGiS) i assotsiirovannykh geneticheskikh simpoziumov (Abstracts of the 6th Congress of the Vavilov Society of Geneticists and Breeders and Associated Genetic Symposiums), Rostov-on-Don, 2014, p. 72.
Dénarié, J., Debellé, F., Promé, J.C., Rhizobium lipochitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis, Annu. Rev. Biochem., 1996, vol. 65, pp. 503–535.
Leppyanen, I.V., Biological synthesis of chitin oligomers and their terminally deacetylated derivatives by enzymes of nodule bacteria, Cand. Sci. (Biol.) Dissertation, St. Petersburg: St. Petersburg. State Univ., 2013.
Madsen, E.B., Madsen, L.H., Radutoiu, S., et al., A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals, Nature, 2003, vol. 425, no. 6958, pp. 637–640.
Radutoiu, S., Madsen, L.H., Madsen, E.B., et al., Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases, Nature, 2003, vol. 425, no. 6958, pp. 585–592.
Zhukov, V., Radutoiu, S., Madsen, L.H., et al., The pea Sym37 receptor kinase gene controls infection-thread initiation and nodule development, Mol. Plant Microbe Interact., 2008, vol. 21, no. 12, pp. 1600–1608.
Kirienko, A.N., Porozov, Yu.B., and Dolgikh, E.A., The role of the new LysM-receptor-like kinase K1 of pea in the development of symbiosis with nodule bacteria, in Biologiya—nauka XXI veka (Biology—The Science of 21th Century) (Collection of Abstracts of 17th Int. Pushchinskaya School–Conf. Young Sci.), Pushchino, 2013, p. 199.
Razumovskaya, Z.G., Nodulation in different pea cultivars, Mikrobiologiya, 1937, vol. 6, no. 3, pp. 321–328.
Lie, T.A., Symbiotic specialization in pea plants: the requirement of specific Rhizobium strains for peas from Afghanistan, Ann. Appl. Biol., 1978, vol. 88, pp. 462–465.
Firmin, J.L., Wilson, K.E., Carlson, R.W., et al., Resistance to nodulation of cv. Afghanistan peas is overcome by nodX, which mediates an O-acetylation of the Rhizobium leguminosarum lipo-oligosaccharide nodulation factor, Mol. Microbiol., 1993, vol. 10, no. 2, pp. 351–360.
Zhukov, V.A., Rychagova, T.S., Shtark, O.Yu., et al., The genetic control of specificity of interactions between legume plants and nodule bacteria, Ekol. Genet., 2008, vol. 6, no. 4, pp. 12–19.
Zhukov, V.A., Sulima, A.S., Porozov, Y.B., et al., Polymorphism in gene sequence of LysM receptor kinase is associated with Sym2-controlled nodulation in pea (Pisum sativum L.), in Proceedings of 18th International Conference on Nitrogen Fixation, Myazaki, 2013, p. 76.
Limpens, E., Franken, C., Smit, P., et al., LysM domain receptor kinases regulating rhizobial Nod factor-induced infection, Science, 2003, vol. 302, no. 5645, pp. 630–633.
Li, R., Knox, M.R., Edwards, A., et al., Natural variation in host-specific nodulation of pea is associated with a haplotype of the SYM37 LysM-type receptorlike kinase, Mol. Plant Microbe Interact., 2011, vol. 24, no. 11, pp. 1396–1403.
Maillet, F., Poinsot, V., André, O., et al., Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza, Nature, 2011, vol. 469, no. 7328, pp. 58–63.
Parniske, M., Arbuscular mycorrhiza: the mother of plant root endosymbioses, Nat. Rev. Microbiol., 2008, vol. 6, pp. 763–775.
Endre, G., Kereszt, A., Kevei, Z., et al., A receptor kinase gene regulating symbiotic nodule development, Nature, 2002, vol. 417, no. 6892, pp. 962–966.
Stracke, S., Kistner, C., Yoshida, S., et al., A plant receptor-like kinase required for both bacterial and fungal symbiosis, Nature, 2002, vol. 417, no. 6892, pp. 959–962.
Oldroyd, G.E. and Downie, J.A., Calcium, kinases and nodulation signalling in legumes, Nat. Rev. Mol. Cell. Biol., 2004, vol. 5, no. 7, pp. 566–576.
Kouchi, H., Imaizumi-Anraku, H., Hayashi, M., et al., How many peas in a pod? Legume genes responsible for mutualistic symbioses underground, Plant Cell Physiol., 2010, vol. 51, no. 9, pp. 1381–1397.
Yano, K. and Yoshida, S., Müller, J., et al., CYCLOPS, a mediator of symbiotic intracellular accommodation, Proc. Natl. Acad. Sci. U.S.A., 2008, vol. 105, no. 51, pp. 20540–20545.
Messinese, E., Mun, J.H., Yeun, L.H., et al., A novel nuclear protein interacts with the symbiotic DMI3 calcium- and calmodulin-dependent protein kinase of Medicago truncatula, Mol. Plant Microbe Interact., 2007, vol. 20, no. 8, pp. 912–921.
Ovchinnikova, E., Journet, E.P., Chabaud, M., et al., IPD3 controls the formation of nitrogen-fixing symbiosomes in pea and Medicago spp., Mol. Plant Microbe Interact., 2011, vol. 24, no. 11, pp. 1333–1344.
Tirichine, L., Imaizumi-Anraku, H., Yoshida, S., et al., Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development, Nature, 2006, vol. 441, no. 7097, pp. 1153–1156.
Sanchez, L., Weidmann, S., Arnould, C., et al., Pseudomonas fluorescens and Glomus mosseae trigger DMI3-dependent activation of genes related to a signal transduction pathway in roots of Medicago truncatula, Plant Physiol., 2005, vol. 139, no. 2, pp. 1065–1077.
Krasil’nikov, N.A. and Melkumova, T.A., Variation of nodule bacteria in the nodules of leguminous plants, Izv. Akad. Nauk SSSR, 1963, no. 5, pp. 693–706.
Osterman, J., Chizhevskaja, E.P., Andronov, E.E., et al., Galega orientalis is more diverse than Galega officinalis in Caucasus—whole-genome AFLP analysis and phylogenetics of symbiosis-related genes, Mol. Ecol., 2011, vol. 20, no. 22, pp. 4808–4821.
Andronov, E.E., Onishchuk, O.P., Kurchak, O.N., and Provorov, N.A., Population structure of the clover rhizobia Rhizobium leguminosarum bv. trifolii upon transition from soil into the nodular niche, Microbiology, 2014, vol. 83, no. 4, p. 422–429.
Porozov, Yu.B., Muntyan, A.N., Chizhevskaya, E.P., et al., Linked symbiotic populations: analysis of polymorphism in nfr5 receptor gene by using molecular doking, Ekol. Genet., 2012, vol. 10, no. 1, pp. 12–18.
Østerås, M., Boncompagni, E. Vincent, N., et al., Presence of a gene encoding choline sulfatase in Sinorhizobium meliloti bet-operon: choline-O-sulfate is metabolized into glycine betaine, Proc. Natl. Acad. Sci. U.S.A., 1998, vol. 95, no. 19, pp. 11394–11399.
Boscari, A., Mandon, K., Dupont, L., et al., BetS is a major glycine betaine/proline betaine transporter required for early osmotic adjustment in Sinorhizobium meliloti, J. Bacteriol., 2002, vol. 184, no. 10, pp. 2654–2663.
Yurgel, S.N., Rice, J., Mulder, M., et al., Truncated betB2-144 plays a critical role in Sinorhizobium meliloti Rm2011 osmoprotection and glycine–betaine catabolism, Eur. J. Soil Biol., 2013, vol. 54, pp. 48–55.
Rumyantseva, M.L., Belova, V.S., Onishchuk, O.P., et al., Polymorphism of bet genes among Sinorhizobium meliloti isolates native to gene centers of alfalfa, S-kh. Biol., 2011, no. 3, pp. 48–54.
Pitsik, E.V., The study of betS locus of nodule bacteria Sinorhizobium meliloti and its value under salinity conditions, Magister Sci. (Biol.) Dissertation, St. Petersburg: St. Petersburg State Univ., 2008, p. 70.
Finan, T.M., Evolving insights: symbiosis islands and horizontal gene transfer, J. Bacteriol., 2002, vol. 184, no. 11, pp. 2855–2856.
Guo, F.-B., Wei, W., Wang, X.L., et al., Co-evolution of genomic islands and their bacterial hosts revealed through phylogenetic analyses of 17 groups of homologous genomic islands, Genet. Mol. Res., 2012, vol. 11, no. 4, pp. 3735–3743.
Dobrindt, U., Hochhut, B., Hentschel, U., and Hacker, J., Genomic islands in pathogenic and environmental microorganisms, Nat. Rev. Microbiol., 2004, vol. 2, no. 5, pp. 414–424.
Ulvé, V.M., Sevin, E.W., Chéron, A., and Barloy-Hubler, F., Identification of chromosomal alpha-proteobacterial small RNAs by comparative genome analysis and detection in Sinorhizobium meliloti strain 1021, BMC Genomics, 2007, vol. 8, p. 467.
Capela, D., Barloy-Hubler, F., Gouzy, J., et al., Analysis of the chromosome sequence of the legume symbiont Sinorhizobium meliloti, Proc. Natl. Acad. Sci. U.S.A., vol. 98, no. 17, pp. 9877–9882.
Becker, A., Barnett, M.J., Capela, D., et al., A portal for rhizobial genomes: RhizoGATE integrates a Sinorhizobium meliloti genome annotation update with postgenome data, J. Biotechnol., 2009, vol. 140, nos. 1–2, pp. 45–50.
Ferri, L., Gori, A., Biondi, E.G., et al., Plasmid electroporation of Sinorhizobium strains: the role of the restriction gene hsdR in type strain Rm1021, Plasmid, 2010, vol. 63, pp. 128–135.
Becker, A., Bergés, H., Krol, E., et al., Global changes in gene expression in Sinorhizobium meliloti 1021 under microoxic and symbiotic conditions, Mol. Plant Microbe Interact., 2004, vol. 17, no. 3, pp. 292–303.
Krol, E. and Becker, A., Global transcriptional analysis of phosphate stress responses in Sinorhizobium meliloti strains 1021 and 2011, Mol. Gen. Genomics, 2004, vol. 272, no. 1, pp. 1–17.
Domínguez-Ferreras, A., Perez-Arnedo, R., Becker, A., et al., Transcriptome profiling reveals the importance of plasmid pSymB for osmoadaptation of Sinorhizobium meliloti, J. Bacteriol., 2006, vol. 188, no. 21, pp. 7617–7625.
Rüberg S., Tian, Z.-X. Krol, E., et al., Construction and validation of a Sinorhizobium meliloti whole genome DNA microarray: genome-wide profiling of osmoadaptive gene expression, J. Biotechnol., 2003, vol. 106, nos. 2–3, pp. 255–268.
de Weert, S., Dekkers, L.C., Kuiper, I., et al., Generation of enhanced competitive root-tip-colonizing Pseudomonas bacteria through accelerated evolution, J. Bacteriol., 2004, vol. 186, no. 10, pp. 3153–3159.
Madsen, L.H., Tirichine, L., Jurkiewicz, A., et al., The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus, Nat. Commun., 2010, vol. 1, p. 10.
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © I.A. Tikhonovich, E.E. Andronov, A.Yu. Borisov, E.A. Dolgikh, A.I. Zhernakov, V.A. Zhukov, N.A. Provorov, M.L. Roumiantseva, B.V. Simarov, 2015, published in Genetika, 2015, Vol. 51, No. 9, pp. 973–990.
Rights and permissions
About this article
Cite this article
Tikhonovich, I.A., Andronov, E.E., Borisov, A.Y. et al. The principle of genome complementarity in the enhancement of plant adaptive capacities. Russ J Genet 51, 831–846 (2015). https://doi.org/10.1134/S1022795415090124
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795415090124