Abstract
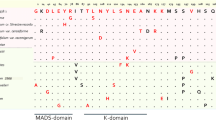
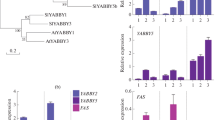
The RIPENING INHIBITOR transcription factor (RIN) is one of the key regulators of the cascade of tomato ripening reactions. RIN triggers many structural and regulatory genes on which such aspects of tomato fruit ripening as the loss of chlorophyll, biosynthesis of carotenoids, aromatic components, organic acids, modification of the structure of cellular walls, and accumulation of sugars depend by controlling both ethylene-dependent and ethylene-independent ripening pathways. In this study, the orthologs of the RIN gene were cloned and sequenced for the first time in seven tomato cultivars (Solanum lycopersicum L.), two wild species used in the breeding process (S. pimpinellifolium L. and S. cheesmaniae (L. Riley) Fosberg), and a wild-growing S. lycopersicum L. It was shown that the polymorphism of the nucleotide and amino acid sequences of RIN in the group of the studied varieties is higher than in the analyzed wild-growing species. The variable sites in the RIN sequence were found to be radical in the majority of cases. The expression profile of two isoforms of the RIN gene, RIN1i and RIN2i, was estimated during fruit ripening in five S. lycopersicum cultivars and a wild species S. cheesmaniae. A positive correlation between the number of RIN2i transcripts and the sugar content in a ripe fruit was found. The expression of both isoforms of the RIN gene was shown to be activated in response to the exogenous action of sucrose. These facts suggest not only the effect of RIN on the accumulation of sugars during ripening but also the opposite effect: an increase in the concentration of sugars leads to an increase in the expression of the RIN gene.




Similar content being viewed by others
REFERENCES
Quinet, M., Angosto, T., Yuste-Lisbona, F.J., Blanchard-Gros, R., Bigot, S., Martinez, J.P., and Lutts, S., Tomato fruit development and metabolism, Front. Plant Sci., 2019, vol. 10, p. 1554. https://doi.org/10.3389/fpls.2019.01554
Karlova, R., Chapman, N., David, K., Angenent, G.C., Seymour, G.B., and de Maagd, R.A., Transcriptional control of fleshy fruit development and ripening, J. Exp. Bot., 2014, vol. 65, p. 4527. https://doi.org/10.1093/jxb/eru316
Li, S., Chen, K., and Grierson, D., A critical evaluation of the role of ethylene and MADS transcription factors in the network controlling fleshy fruit ripening, New Phytol., 2019, vol. 221, p. 1724. https://doi.org/10.1111/nph.15545
Robinson, R. and Tomes, M., Ripening inhibitor: a gene with multiple effect on ripening tomato, Genet. Coop., 1968, vol. 18, p. 36.
Tigchelaar, E., Tomes, M., Kerr, E., and Barman, R., A new fruit ripening mutant, non-ripening (nor), Rep. Tomato Genet. Coop., 1973, vol. 23, p. 33.
Thompson, A.J., Tor, M., Barry, C.S., Vrebalov, J., Orfila, C., Jarvis, M.C., Giovannoni, J.J., Grierson, D., and Seymour, G.B., Molecular and genetic characterization of a novel pleiotropic tomato-ripening mutant, Plant Physiol., 1999, vol. 120, p. 383.
Zhong, S., Fei, Z., Chen, Y.R., Zheng, Y., Huang, M., Vrebalov, J., McQuinn, R., Gapper, N., Liu, B., Xiang, J., Shao, Y., and Giovannoni, J.J., Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening, Nat. Biotechnol., 2013, vol. 31, p. 154.
Vrebalov, J., Ruezinsky, D., Padmanabhan, V., White, R., Medrano, D., Drake, R., Schuch, W., and Giovannoni, J., A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus, Science, 2002, vol. 296, p. 343.
Fujisawa, M., Nakano, T., Shima, Y., and Ito, Y., A large-scale identification of direct targets of the tomato MADS box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening, Plant Cell, 2013, vol. 25, p. 371.
Qin, G., Wang, Y., Cao, B., Wang, W., and Tian, S., Unraveling the regulatory network of the MADS box transcription factor RIN in fruit ripening, Plant J., 2012, vol. 70, p. 243.
Osorio, S., Alba, R., Damasceno, C.M., Lopez-Casado, G., Lohse, M., Zanor, M.I., Tohge, T., Usadel, B., Rose, J.K., Fei, Z., Giovannoni, J.J., and Fernie, A.R., Systems biology of tomato fruit development: combined transcript, protein, and metabolite analysis of tomato transcription factor (nor, rin) and ethylene receptor (Nr) mutants reveals novel regulatory interactions, Plant Physiol., 2011, vol. 157, p. 405.
Ito, Y., Regulation of tomato fruit ripening by MADS-box transcription factors, Horticulture, 2016, vol. 50, p. 33.
Bondareva, L.L., Metodicheskie ukazaniya po aprobatsii oboshchnykh i bakhchevykh kul’tur (Guide for Test Cultivation of Vegetable and Melon Cultures), Moscow: Fed. Nauchn. Tsentr Ovoshchevod., 2018.
Bhattarai, K., Sharma, S., and Panthee, D.R., Diversity among modern tomato genotypes at different levels in fresh-market breeding, Int. J. Agron., 2018, vol. 2018, p. 4170432. https://doi.org/10.1155/2018/4170432
Kang, S.I., Hwang, I., Goswami, G., Jung, H.J., Nath, U.K., Yoo, H.J., Lee, J.M., and Nou, I.S., Molecular insights reveal Psy1, SGR, and SlMYB12 genes are associated with diverse fruit color pigments in tomato (Solanum lycopersicum L.), Molecules, 2017, vol. 22, p. 2180. https://doi.org/10.3390/molecules22122180
D'Angelo, M., Zanor, M.I., Sance, M., Cortina, P.R., Boggio, S.B., Asprelli, P., Carrari, F., Santiago, A.N., Asís, R., Peralta, I.E., and Valle, E.M., Contrasting metabolic profiles of tasty Andean varieties of tomato fruit in comparison with commercial ones, J. Sci. Food Agric., 2018, vol. 98, p. 4128. https://doi.org/10.1002/jsfa.8930
Pratta, G.R., Rodriguez, G.R., Zorzoli, R., Valle, E.M., and Picardi, L.A., Phenotypic and molecular characterization of selected tomato recombinant inbred lines derived from the cross Solanum lycopersicum × S. pimpinellifolium, J. Genet., 2011, vol. 90, p. 229. https://doi.org/10.1007/s12041-011-0063-0
Edwards, K., Johnstone, C., and Thompson, C., A simple and rapid method for the preparation of plant genomic DNA for PCR analysis, Nucleic Acids Res., 1991, vol. 19, p. 1349. https://doi.org/10.1093/nar/19.6.1349
Efremov, G.I., Slugina, M.A., Shchennikova, A.V., and Kochieva, E.Z., Differential regulation of phytoene synthase PSY1 during fruit carotenogenesis in cultivated and wild tomato species (Solanum section Lycopersicon), Plants, 2020, vol. 9, p. 1169. https://doi.org/10.3390/plants9091169
Filyushin, M.A., Dzhos, E.A., Shchennikova, A.V., and Kochieva, E.Z., Dependence of pepper fruit color on basic pigments ratio and expression pattern of carotenoid and anthocyanin biosynthesis genes, Russ. J. Plant Physiol., 2020, vol. 67, p. 1054. https://doi.org/10.1134/S1021443720050040
Henareh, M., Dursun, A., and Mandoulakani, B.A., Genetic diversity in tomato landraces collected from Turkey and Iran revealed by morphological characters, Acta Sci. Pol., Hortorum Cult., 2015, vol. 14, p. 87.
Cebolla-Cornejo, J., Roselló, S., and Nuez, F., Phenotypic and genetic diversity of Spanish tomato landraces, Sci. Hortic. (Amsterdam), 2013, vol. 162, p. 150.
Lin, T., Zhu, G., Zhang, J., Xu, X., Yu, Q., Zheng, Z., Zhang, Z., Lun, Y., Li, S., Wang, X., Huang, Z., Li, J., Zhang, C., Wang, T., Zhang, Y., et al., Genomic analyses provide insights into the history of tomato breeding, Nat. Genet., 2014, vol. 46, p. 1220. https://doi.org/10.1038/ng.3117
Aerts, N., de Bruijn, S., van Mourik, H., Angenent, G.C., and van Dijk, A.D.J., Comparative analysis of binding patterns of MADS-domain proteins in Arabidopsis thaliana, BMC Plant Biol., 2018, vol. 18, p. 131. https://doi.org/10.1186/s12870-018-1348-8
Kaufmann, K., Melzer, R., and Theissen, G., MIKC-type MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants, Gene, 2005, vol. 347, p. 183. https://doi.org/10.1016/j.gene.2004.12.014
Immink, R.G., Tonaco, I.A., de Folter, S., Shchennikova, A., van Dijk, A.D., Busscher-Lange, J., Borst, J.W., and Angenent, G.C., SEPALLATA3: the ‘glue’ for MADS box transcription factor complex formation, Genome Biol., 2009, vol. 10, p. R24. https://doi.org/10.1186/gb-2009-10-2-r24
Qin, G., Zhu, Z., Wang, W., Cai, J., Chen, Y., Li, L., and Tian, S., A tomato vacuolar invertase inhibitor mediates sucrose metabolism and influences fruit ripening, Plant Physiol., 2016, vol. 172, p. 1596. https://doi.org/10.1104/pp.16.01269
Ito, Y., Sekiyama, Y., Nakayama, H., Nishizawa-Yokoi, A., Endo, M., Shima, Y., Nakamura, N., Kotake-Nara, E., Kawasaki, S., Hirose, S., and Toki, S., Allelic mutations in the ripening-inhibitor (RIN) locus generate extensive variation in tomato ripening, Plant Physiol., 2020, vol. 183, p. 80. https://doi.org/10.1104/pp.20.00020
Rolland, F., Moore, B., and Sheen, J., Sugar sensing and signaling in plants, Plant Cell, 2002, vol. 14, p. S185. https://doi.org/10.1105/tpc.010455
Beaujean, A., Ducrocq-Assaf, C., Sangwan, R.S., Lilius, G., Bülow, L., and Sangwan-Norreel, B.S., Engineering direct fructose production in processed potato tubers by expressing a bifunctional alpha-amylase/glucose isomerase gene complex, Biotechnol. Bioeng., 2000, vol. 70, p. 9. https://doi.org/10.1002/1097-0290(20001005)70:1<9::aid-bit2>3.0.co;2-7
Funding
This work was supported by a grant from the Russian Science Foundation, no. 19-76-00006, and, in part, from the Ministry of Science and Higher Education of the Russian Federation, using an experimental artificial climate setup (EACS, Federal Research Center for Biotechnology, Russian Academy of Sciences).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests. The authors declare that they have no conflicts of interest.
Statement on the welfare of humans or animals. This article does not contain any studies involving animals performed by any of the authors.
Additional information
Translated by M. Shulskaya
Supplementary Information
Rights and permissions
About this article
Cite this article
Slugina, M.A., Dzhos, E.A., Schennikova, A.V. et al. The Sugar Content in the Tomato Ripe Fruit Correlates with the Expression Level of the RIN2i Isform of the Ripening Inhibitor Gene. Russ J Plant Physiol 68, 1038–1047 (2021). https://doi.org/10.1134/S1021443721050198
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1021443721050198