Abstract
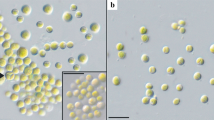
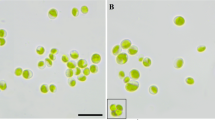
The species diversity of green coccoid algae is often difficult to study using light microscopy due to their complex morphology. Algae, which have morphological and ecological similarities with the genera Bracteacoccus Tereg., Dictyococcus Gerneck, and Pseudomuriella Hanagata, are one of the most taxonomically complex examples. Often, only a comparison of nucleotide sequences allows one to carry out a reliable identification and find out the phylogenetic identity of the studied objects. Pseudomuriella belongs to the group of genetically diverse genera with a homogeneous Bracteacoccus-like morphology. The combined use of two genetic markers (18S rDNA nuclear gene and chloroplast rbcL gene), along with light microscopy and comparison of fatty acid profiles, allowed the identification of the green algae strain isolated from the soil of the artificial pine and robinia plantation in the steppe zone of Ukraine as Pseudomuriella engadinensis (Kol et Chodat) Fučiková, Rada et Lewis—the first find of the species in the flora of algae in Ukraine. Analysis of the fatty acid composition of the studied strain’s cells showed that the total lipid content in the stationary growth phase was at the level of 87.9 ± 2.1 mg/g dry cell mass, and the main fatty acids were palmitic, hexadecadienoic, ruganic, oleic, linoleic, and α-linolenic—their share accounted for 82.4% of the total amount of fatty acids.



Similar content being viewed by others
REFERENCES
Maltsev, Y.I., Konovalenko, T.V., Barantsova, I.A., Maltseva, I.A., and Maltseva, K.I., Prospects of using algae in biofuel production, Regul. Mech. Biosyst., 2017, vol. 8, pp. 455–460. https://doi.org/10.15421/021770
Mudimu, O., Koopmann, I.K., Rybalka, N., Friedl, T., Schulz, R., and Bilger, W., Screening of microalgae and cyanobacteria strains for α-tocopherol content at different growth phases and the influence of nitrate reduction on α-tocopherol production, J. Appl. Phycol., 2017, vol. 29, pp. 2867–2875. https://doi.org/10.1007/s10811-017-1188-1
Fučiková, K., Rada, J.C., and Lewis, L.A., The tangled taxonomic history of Dictyococcus, Bracteacoccus and Pseudomuriella (Chlorophyceae, Chlorophyta) and their distinction based on a phylogenetic perspective, Phycologia, 2011, vol. 50, pp. 422–429. https://doi.org/10.2216/10-69.1
Fučiková, K. and Lewis, L.A., Intersection of Chlorella, Muriella and Bracteacoccus: resurrecting the genus Chromochloris Kol et Chodat (Chlorophyceae, Chlorophyta), Fottea, 2012, vol. 12, pp. 83–93.
Hanagata, N., Phylogeny of the subfamily Scotiellocystoideae (Chlorophyceae, Chlorophyta) and related taxa inferred from 18S ribosomal RNA gene sequence data, J. Phycol., 1998, vol. 34, pp. 1049–1054.
Fučiková, K., Rada, J.C., Lukešová, A., and Lewis, L.A., Cryptic diversity within the genus Pseudomuriella Hanagata (Chlorophyta, Chlorophyceae, Sphaeropleales) assessed using four Barcode markers, Nova Hedw., 2011, vol. 93, pp. 29–46. https://doi.org/10.1127/0029-5035/2011/0093-0029
Fučiková, K., Lewis, P.O., and Lewis, L.A., Putting incertae sedis taxa in their place: a proposal for ten new families and three new genera in Sphaeropleales (Chlorophyceae, Chlorophyta), J. Phycol., 2014, vol. 50, pp. 14–25. https://doi.org/10.1111/jpy.12118
Kalina, T. and Punčochářová, M., Taxonomy of the subfamily Scotiellocystoideae Fott 1976 (Chlorellaceae, Chlorophyceae), Arch. Hydrobiol., 1987, vol. 73, pp. 473–521.
Caisová, L. and Melkonian, M., Evolution of helix formation in the ribosomal internal transcribed spacer 2 (ITS2) and its significance for RNA secondary structures, J. Mol. Evol., 2014, vol. 78, pp. 324–337. https://doi.org/10.1007/s00239-014-9625-0
Mikhailyuk, T., Kondratyuk, S.Ya., Niporko, S.O., Darienko, T.M., Demchenko, E.M., and Voitsekhovich, A.O., Lishainiki, mohopodobnii i nazemnii vodorosli granitnih kan’onov Ukraini (Lichens, Mosses, and Terrestrial Algae of Granite Canyons of Ukraine), Kiev: Alterpress, 2011.
Bischoff, H.W. and Bold, H.C., Bracteacoccus grandis, in Phycological Studies IV. Some Soil Algae from Enchanted Rock and Related Algal Species, University of Texas Press, 1963, publ. no. 6318.
Zimmermann, J., Jahn, R., and Gemeinholzer, B., Barcoding diatoms: evaluation of the V4 subregion on the 18S rRNA gene, including new primers and protocols, Org. Divers. Evol., 2011, vol. 11: 173. https://doi.org/10.1007/s13127-011-0050-6
Nakada, T., Shinkawa, H., Ito, T., and Tomita, M., Recharacterization of Chlamydomonas reinhardtii and its relatives with new isolates from Japan, J. Plant Res., 2010, vol. 123, pp. 67–78.
Katoh, K. and Toh, H., Parallelization of the MAFFT multiple sequence alignment program, Bioinformatics, 2010, vol. 26, pp. 1899–1900.
Stamatakis, A., Hoover, P., and Rougemont, J., A rapid bootstrap algorithm for the RAxML web-servers, Syst. Biol., 2008, vol. 75, pp. 758–771.
Ronquist, F. and Huelsenbeck, J.P., MrBayes 3: Bayesian phylogenetic inference under mixed models, Bioinformatics, 2003, vol. 19, pp. 1572–1574.
Starikov, A.Yu., Userbaeva, A.A., Mironov, K.S., Sidorov, R.A., Zayadan, B.K., Bedbenov, V.S., Sinetova, M.A., and Los, D.A., Substrate specificity of acyl-lipid [delta]9-desaturase from Cyanobacterium sp. IPPAS B-1200, a Cyanobacterium with unique fatty acid composition, Russ. J. Plant Physiol., 2018, vol. 65, pp. 490–497.
Neustupa, J.I., Chlorophyta, Streptophyta p.p. (except Ulvophyceae, Charophyceae; incl. Trentepohliales), in Syllabus of Plant Families. A. Engler’s Syllabus der Pflanzenfamilien, Part 2/1: Photoautotrophic Eukaryotic Algae Glaucocystophyta, Cryptophyta, Dinophyta/Dinozoa, Haptophyta, Heterokontophyta/Ochrophyta, Chlorarachniophyta/Cercozoa, Euglenophyta/Euglenozoa, Chlorophyta, Streptophyta p.p., Wolfgang, F., Ed., Schweizerbart/Borntraeger Sci. Publ., 2015, pp. 190–247.
Kol, E. and Chodat, F., Quelques algues nouvelles des sols et de la neige du Parc National Suisse, Engadine, Bull. Soc. Bot. Genève, 1934, vol. 25, pp. 250–263.
Starr, R.C., A Comparative Study of Chlorococcum Meneghini and Other Spherical, Zoospore Producing Genera of the Chlorococcales, Bloomington: Indiana Univ. Press, 1955.
Maltsev, Y. and Kulikovskiy, M., Molecular and morphological investigation of cosmopolitan diatom Hantzschia amphioxys with remarks on biogeography, Phycologia, 2017, vol. 56, no. 4: 125.
Kostikov, I.Yu., Romanenko, P.O., Demchenko, E.M., Darienko, T.M., Mikhailyuk, T.I., Ribchins’kii, O.V., and Solonenko, A.M., Vodorosli gruntov Ukraini: istoriya i metody issledovaniya, sistema konspektflory (Soil Algae of Ukraine: History and Methods of Research, System of Floral Outline), Kiev: Fitosociocenter, 2001.
Maltseva, I.A., Gruntovi vodorosti lisiv stepovoyi zony Ukrayiny (Soil Algae of the Steppe Zone of Ukraine), Melitopol’: Lyuks, 2009.
Scherbina, V.V., Maltseva, I.A., and Solonenko, A.N., Peculiarities of postpyrogene development of algae in steppe biocenoses at Askania Nova Biospheric National Park, Contemp. Probl. Ecol., 2014, vol. 7, pp. 187–191. https://doi.org/10.1134/S1995425514020140
Maltsev, Y.I., Pakhomov, A.Y., and Maltseva, I.A., Specific features of algal communities in forest litter of forest biogeocenoses of the steppe zone, Contemp. Probl. Ecol., 2017, vol. 10, pp. 71–76. https://doi.org/10.1134/S1995425517010085
Maltsev, Y.I., Didovich, S.V., and Maltseva, I.A., Seasonal changes in the communities of microorganisms and algae in the litters of tree plantations in the steppe zone, Eurasian Soil Sci., 2017, vol. 50, pp. 935–942. https://doi.org/10.1134/S1064229317060059
Maltsev, Y.I., Maltseva, I.A., Solonenko, A.N., and Bren, A.G., Use of soil biota in the assessment of the ecological potential of urban soils, Biosyst. Divers., 2017, vol. 25, pp. 257–262. https://doi.org/10.15421/011739
Algae of Ukraine: Diversity, Nomenclature, Taxonomy, Ecology and Geography, Vol. 3: Chlorophyta, Tsarenko, P.M., Wasser, S., and Nevo, E., Eds., Gantner Verlag, 2011.
Vinogradova, O.N. and Darienko, T.M., Algae of Azovo-Syvashsky National Nature Park (Ukraine), Int. J. Algae, 2008, vol. 10, pp. 163–178. https://doi.org/10.1615/InterJAlgae.v10.i2.50
Lang, I., Hodac, L., Friedl, T., and Feussner, I., Fatty acid profiles and their distribution patterns in microalgae: a comprehensive analysis of more than 2000 strains from the SAG culture collection, BMC Plant Biol., 2011, vol. 11: 124. https://doi.org/10.1186/1471-2229-11-124
Funding
This publication is based on research carried out with financial support by Russian Science Foundation (project number 18-74-00095).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
Additional information
Translated by M. Shulskaya
Abbreviations: BI—Bayesian inference; DO—directly opposed orientation of the basal bodies of flagella, phylogenetic clade of the same name; FAME—fatty acid methyl esters; FFA—free fatty acids; LB—likelihood bootstrap; ML—maximum likelihood; PP—posterior probability.
Supplementary material
Rights and permissions
About this article
Cite this article
Maltsev, Y.I., Maltseva, I.A., Kulikovskiy, M.S. et al. Analysis of a New Strain of Pseudomuriella engadinensis (Sphaeropleales, Chlorophyta) for Possible Use in Biotechnology. Russ J Plant Physiol 66, 609–617 (2019). https://doi.org/10.1134/S1021443719040083
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1021443719040083