Abstract
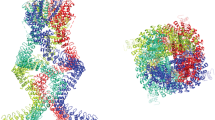
The marine free-living organism Trichoplax (phylum Placozoa) resembles a unicellular amoeba in shape and type of movement. Trichoplax diverged from the main evolutionary tree in the Neoproterozoic Era. Trichoplax provides one of the simplest models of multicellular animals and a strong example of how cells of an organism interact to form an ensemble during its development and movement. Two orthologs of the mouse Piezo1 protein (6B3R) were found in two Trichoplax haplotypes, H1 and H2, as a result of a search for similar sequences in the NCBI databases. Spatial models of the respective proteins XP_002112008.1 and RDD46920.1 were created via a structural alignment with 6KG7 (mouse Piezo2) template. Their domain structures were analyzed, and a limited graph of protein‒protein interactions was constructed for the hypothetical mechanosensor XP_002112008.1. The possibility of signal transduction from the mechanoreceptor to membrane complexes, the cytoplasm, and the cell nucleus was shown. Trichoplax mechanoreceptors were assumed to play a role in perception of force stimuli from neighbor cells and the environment. Based on the results, the primitive Trichoplax organism was proposed as the simplest multicellular model of mechanical and morphogenetic movements.


Similar content being viewed by others
REFERENCES
Niethammer P. 2021. Components and mechanisms of nuclear mechanotransduction. Annu. Rev. Cell Dev. Biol. 37, 233‒256. https://doi.org/10.1146/annurev-cellbio-120319-030049
Fajardo-Cavazos P., Nicholson W.L. 2021. Mechanotransduction in prokaryotes: A possible mechanism of spaceflight adaptation. Life (Basel). 11 (1), 33. https://doi.org/10.3390/life11010033
Jin P., Jan L.Y., Jan Y.N. 2020. Mechanosensitive ion channels: Structural features relevant to mechanotransduction mechanisms. Annu. Rev. Neurosci. 43, 207‒229. https://doi.org/10.1146/annurev-neuro-070918-050509
Marshall K.L., Lumpkin E.A. 2012. The molecular basis of mechanosensory transduction. Adv Exp. Med. Biol. 739, 142‒155. https://doi.org/10.1007/978-1-4614-1704-0_9
Clapham D.E. 2007. Calcium signaling. Cell. 131 (6), 1047‒1058. https://doi.org/10.1016/j.cell.2007.11.028
Perozo E. 2006. Gating prokaryotic mechanosensitive channels. Nat. Rev. Mol. Cell Biol. 7 (2), 109‒119. https://doi.org/10.1038/nrm1833
Arnadóttir J., Chalfie M. 2010. Eukaryotic mechanosensitive channels. Annu. Rev. Biophys. 39, 111‒137. https://doi.org/10.1146/annurev.biophys.37.032807.125836
Earley S., Santana L.F., Lederer W.J. 2021. The physiological sensor channels TRP and Piezo: Nobel prize in physiology or medicine. Physiol. Rev. 102 (2), 1153‒1158. https://doi.org/10.1152/physrev.00057.2021
Du G., Chen W., Li L., Zhang Q. 2022. The potential role of mechanosensitive ion channels in substrate stiffness-regulated Ca2+ response in chondrocytes. Connect. Tissue Res. 63 (5), 453‒462. https://doi.org/10.1080/03008207.2021.2007902
Coste B., Mathur J., Schmidt M., Earley T.J., Ranade S., Petrus M.J., Dubin A.E., Patapoutian A. 2010. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science. 330 (6000), 55‒60. https://doi.org/10.1126/science.1193270
Fang X.Z., Zhou T., Xu J.Q., Wang Y.X., Sun M.M., He Y.J., Pan S.W., Xiong W., Peng Z.K., Gao X.H., Shang Y. 2021. Structure, kinetic properties and biological function of mechanosensitive Piezo channels. Cell Biosci. 11 (1), 13. https://doi.org/10.1186/s13578-020-00522-z
Barzegari A., Omidi Y., Ostadrahimi A., Gueguen V., Meddahi-Pellé A., Nouri M., Pavon-Djavid G. 2020. The role of Piezo proteins and cellular mechanosensing in tuning the fate of transplanted stem cells. Cell Tissue Res. 381 (1), 1‒12. https://doi.org/10.1007/s00441-020-03191-z
Ge J., Li W., Zhao Q., Li N., Chen M., Zhi P., Li R., Gao N., Xiao B., Yang M. 2015. Architecture of the mammalian mechanosensitive Piezo1 channel. Nature. 527 (7576), 64‒69. https://doi.org/10.1038/nature15247
Coste B., Xiao B., Santos J.S., Syeda R., Grandl J., Spencer K.S., Kim S.E., Schmidt M., Mathur J., Dubin A.E., Montal M., Patapoutian A. 2012. Piezo proteins are pore-forming subunits of mechanically activated channels. Nature. 483 (7388), 176‒181. https://doi.org/10.1038/nature10812
Coste B., Murthy S.E., Mathur J., Schmidt M., Mechioukhi Y., Delmas P., Patapoutian A. 2015. Piezo1 ion channel pore properties are dictated by C-terminal region. Nat. Commun. 6, 7223. https://doi.org/10.1038/ncomms8223
Syeda R., Florendo M.N., Cox C.D., Kefauver J.M., Santos J.S., Martinac B., Patapoutian A. 2016. Piezo1 channels are inherently mechanosensitive. Cell Rep. 17 (7), 1739‒1746. https://doi.org/10.1016/j.celrep.2016.10.033
Syed T., Schierwater B. 2002. The evolution of the Placozoa: A new morphological model. Palaeobiodiversity Palaeoenviron. 82 (1), 315‒324. https://doi.org/10.1007/BF03043791
Srivastava M., Begovic E., Chapman J., Putnam N.H., Hellsten U., Kawashima T., Kuo A., Mitros T., Sa-lamov A., Carpenter M.L., Signorovitch A.Y., Moreno M.A., Kamm K., Grimwood J., Schmutz J., Shapiro H., Grigoriev I.V., Buss L.W., Schierwater B., Dellaporta S.L., Rokhsar D.S. 2008. The Trichoplax genome and the nature of placozoans. Nature. 454 (7207), 955‒960. https://doi.org/10.1038/nature07191
Kamm K., Osigus H.J., Stadler P.F., DeSalle R., Schierwater B. 2018. Trichoplax genomes reveal profound admixture and suggest stable wild populations without bisexual reproduction. Sci. Rep. 8 (1), 11168. https://doi.org/10.1038/s41598-018-29400-y
Smith C.L., Varoqueaux F., Kittelmann M., Azzam R.N., Cooper B., Winters C.A., Eitel M., Fasshauer D., Reese T.S. 2014. Novel cell types, neurosecretory cells, and body plan of the early-diverging metazoan Trichoplax adhaerens. Curr. Biol. 24 (14), 1565‒1572. https://doi.org/10.1016/j.cub.2014.05.046
Wenderoth H. 1990. Cytoplasmic vibrations due to flagellar beating in Trichoplax adhaerens F. E. Schulze (Placozoa). Z. Naturforsch. 45, 715‒722. https://doi.org/10.1515/znc-1990-0624
Armon S., Bull M.S., Aranda-Diaz A., Prakash M. 2018. Ultrafast epithelial contractions provide insights into contraction speed limits and tissue integrity. Proc. Natl. Acad. Sci. U. S. A. 115 (44), E10333‒E10341. https://doi.org/10.1073/pnas.1802934115
Kuznetsov A.V., Halaimova A.V., Ufimtseva M.A., Chelebieva E.S. 2020. Blocking a chemical communication between Trichoplax organisms leads to their disorderly movement. Int. J. Parallel Emergent Distrib. Syst. 35 (4), 473‒482. https://doi.org/10.1080/17445760.2020.1753188
Kuznetsov A.V., Vainer V.I., Volkova Y.M., Kartashov L.E. 2021. Motility disorders and disintegration into separate cells of Trichoplax sp. H2 in the presence of Zn2+ ions and L-cysteine molecules: A systems approach. Biosystems. 206, 104444. https://doi.org/10.1016/j.biosystems.2021.104444
Ueda T., Koya S., Maruyama Y.K. 1999. Dynamic patterns in the locomotion and feeding behaviors by the placozoan Trichoplax adhaerence. Biosystems. 54 (1-2), 65‒70. https://doi.org/10.1016/s0303-2647(99)00066-0
Smith C.L., Reese T.S., Govezensky T., Barrio R.A. 2019. Coherent directed movement toward food modeled in Trichoplax, a ciliated animal lacking a nervous system. Proc. Natl. Acad. Sci. U. S. A. 116 (18), 8901‒8908. https://doi.org/10.1073/pnas.1815655116
Velankar S., Burley S.K., Kurisu G., Hoch J.C., Markley J.L. 2021. The protein data bank archive. Methods Mol. Biol. 2305, 3‒21. https://doi.org/10.1007/978-1-0716-1406-8_1
Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215 (3), 403‒410. https://doi.org/10.1016/S0022-2836(05)80360-2
Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10 (6), 845‒858. https://doi.org/10.1038/nprot.2015.053
Sayle R.A., Milner-White E.J. 1995. RASMOL: Biomolecular graphics for all. Trends Biochem. Sci. 20 (9), 374. https://doi.org/10.1016/s0968-0004(00)89080-5
Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G.A., Sonnhammer E.L.L., Tosatto S.C.E., Paladin L., Raj S., Richardson L.J., Finn R.D., Bateman A. 2021. Pfam: The protein families database in 2021. Nucleic Acids Res. 49 (D1), D412‒D419. https://doi.org/10.1093/nar/gkaa913
Szklarczyk D., Gable A.L., Nastou K.C., Lyon D., Kirsch R., Pyysalo S., Doncheva N.T., Legeay M., Fang T., Bork P., Jensen L.J., von Mering C. 2021. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 49 (D1), D605‒D612. https://doi.org/10.1093/nar/gkaa1074
Guo Y.R., MacKinnon R. 2017. Structure-based membrane dome mechanism for Piezo mechanosensitivity. Elife. 6, e33660. https://doi.org/10.7554/eLife.33660
Wang L., Zhou H., Zhang M., Liu W., Deng T., Zhao Q., Li Y., Lei J., Li X., Xiao B. 2019. Structure and mechanogating of the mammalian tactile channel PIEZO2. Nature. 573 (7773), 225‒229. https://doi.org/10.1038/s41586-019-1505-8
Grigorov M.G. 2005. Global properties of biological networks. Drug Discovery Today. 10 (5), 365‒372. https://doi.org/10.1016/S1359-6446(05)03369-6
Ranade S.S., Syeda R., Patapoutian A. 2015. Mechanically activated ion channels. Neuron. 87 (6), 1162‒1179. https://doi.org/10.1016/j.neuron.2015.08.032
Fadeeva M.V., Kurchenko V.M., Kuznetsov A.V. 2022. Modern trends in biological physics and chemistry BPPC–Proceedings of XVII International Scientific Conference, 2022. Sevastopol, Russia, Abstract book. pp. 201‒202.
Fadeeva M.V., Sergeeva E.V., Rybakova K.A., Kuznetsov A.V. 2022. Characteristics of the cationic TRPA1-channals family in Trichoplax sp. H2 (Placozoa). Russ. J. Biol. Phys. Chem. 7 (3), 493‒450. doi.org/https://doi.org/10.29039/rusjbpc.2022.0550
Scheres B., van der Putten W.H. 2017. The plant perceptron connects environment to development. Nature. 543 (7645), 337‒345. https://doi.org/10.1038/nature22010
Timsit Y., Grégoire S.P. 2021. Towards the idea of molecular brains. Int. J. Mol. Sci. 22 (21), 11868. https://doi.org/10.3390/ijms222111868
Cox C.D., Bavi N., Martinac B. 2019. Biophysical principles of ion-channel-mediated mechanosensory transduction. Cell Rep. 29 (1), 1‒12. https://doi.org/10.1016/j.celrep.2019.08.075
Lewis A.H., Grandl J. 2021. Piezo1 ion channels inherently function as independent mechanotransducers. Elife. 10, e70988. https://doi.org/10.7554/eLife.70988
Yang X., Lin C., Chen X., Li S., Li X., Xiao B. 2022. Structure deformation and curvature sensing of PIEZO1 in lipid membranes. Nature. 604 (7905), 377‒383. https://doi.org/10.1038/s41586-022-04574-8
Young M., Lewis A.H., Grandl J. 2022. Physics of mechanotransduction by Piezo ion channels. J. Gen. Physiol. 154 (7), e202113044. https://doi.org/10.1085/jgp.202113044
ACKNOWLEDGMENTS
We are grateful to Prof. Graciela Pavon-Djavid (Laboratoire de Recherche Vasculaire Translationnelle, Institut Galilée, Université Sorbonne Paris Nord, France) for published data, students V.I. Vainer and Yu.M. Volkova for help in Trichoplax studies, postgraduates A.V. Khalaimova and M.N. Ufimtseva, and anonymous reviewers for their helpful advices on improving the manuscript.
Funding
This work was supported by the Government of the Russian Federation in accordance with Rule no. 220 (agreement no. 14.W03.31.0015 dated 28.02.2017).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest. The authors declare that they have no conflicts of interest.
This work does not contain any studies involving animals or human subjects performed by any of the authors.
Supplementary Information
Rights and permissions
About this article
Cite this article
Kuznetsov, A.V., Grishin, I.Y. & Vtyurina, D.N. Spatial Models of Piezo Proteins and Protein‒Protein Interaction Networks in Trichoplax Animals (Placozoa). Mol Biol 57, 905–912 (2023). https://doi.org/10.1134/S0026893323050072
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893323050072