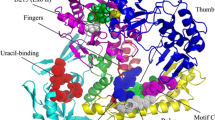
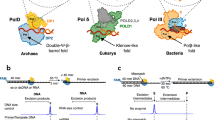
Abstract—DNA polymerases catalyze DNA synthesis during DNA replication, repair, and recombination. A number of DNA polymerases, such as the Taq enzyme from Thermus aquaticus, are used in various applications of molecular biology and biotechnology, in particular as DNA amplification tools. However, the efficiency of these enzymes depends on factors such as DNA origin, primer composition, template length, GC-content, and the ability to form stable secondary structures. These limitations in the use of currently known DNA polymerases lead to the search for new enzymes with improved properties. This review summarizes the main structural and molecular-kinetic features of the functioning of DNA-polymerases belonging to structural family A, including Taq polymerase. A phylogenetic analysis of these enzymes was carried out, which made it possible to establish a highly conserved consensus sequence containing 62 amino acid residues distributed over the structure of the enzyme. A comparative analysis of these amino acid residues among poorly studied DNA-polymerases revealed 7 enzymes that potentially have the properties necessary for use in DNA amplification.






Similar content being viewed by others
REFERENCES
Nikoomanzar A., Chim N., Yik E.J., Chaput J.C. 2020. Engineering polymerases for applications in synthetic biology. Q. Rev. Biophys. 53, 1–31.
Wu D.A.N.Y., Ugozzoli L., Pal B.K., Qian J.I.N., Wallace R.B. 1991. The effect of temperature and oligonucleotide primer length on the specificity and efficiency of amplification by the polymerase chain reaction. DNA Cell Biol. 10, 233–238.
Owczarzy R., Moreira B.G., You Y., Behlke M.A., Walder J.A. 2008. Predicting stability of DNA duplexes in solutions containing magnesium and monovalent cations. Biochemistry. 47, 5336–5353.
Garcia-Diaz M., Bebenek K. 2007. Multiple functions of DNA polymerases. CRC. Crit. Rev. Plant Sci. 26, 105–122.
Alba M.M. 2001. Replicative DNA polymerases. Genome Biol. 2, 1–7.
Rothwell P.J., Waksman G. 2005. Structure and mechanism of DNA polymerases. Adv. Protein Chem. 71, 401–440.
Chien A., Edgar D.B., Trela J.M. 1976. Deoxyribonucleic acid polymerase from the extreme thermophile Thermus aquaticus. J. Bacteriol. 127, 1550–1557.
Choi J.J., Jung S.E., Kim H.K., Kwon S.T. 1999. Purification and properties of Thermus filiformis DNA polymerase expressed in Escherichia coli. Biotechnol. Appl. Biochem. 30, 19–25.
Lawyer F.C., Stoffel S., Saiki R.K., Chang S.Y., Landre P.A., Abramson R.D., Gelfand D.H. 1993. High-level expression, purification, and enzymatic characterization of full-length Thermus aquaticus DNA polymerase and a truncated form deficient in 5′ to 3′ exonuclease activity. Genome Res. 2, 275–287.
Park J.H., Kim J.S., Kwon S.-T., Lee D.-S. 1993. Purification and characterization of Thermus caldophilus GK24 DNA polymerase. Eur. J. Biochem. 214, 135–140.
Kaledin A.S., Sliusarenko A.G., Gorodetskiĭ S.I. 1980. Isolation and properties of DNA polymerase from extreme thermophylic bacteria Thermus aquaticus YT-1. Biokhimiia. 45, 644–651.
Saiki R.K., Gelfand D.H., Stoffel S., Scharf S.J., Higuchi R., Horn G.T., Mullis K.B., Erlich H.A. 1988. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 239, 487–491.
Arezi B., Xing W., Sorge J.A., Hogrefe H.H. 2003. Amplification efficiency of thermostable DNA polymerases. Anal. Biochem. 321, 226–235.
Al-Soud W.A., Rådström P. 2001. Purification and characterization of PCR-inhibitory components in blood cells. J. Clin. Microbiol. 39, 485–493.
Flaman J.-M., Frebourg T., Moreau V., Charbonnier F., Martin C., Ishioka C., Friend S.H., Iggo R. 1994. A rapid PCR fidelity assay. Nucleic Acids Res. 22, 3259–3260.
Ling L.L., Keohavong P., Dias C., Thilly W.G. 1991. Optimization of the polymerase chain reaction with regard to fidelity: modified T7, Taq, and Vent DNA polymerases. Genome Res. 1, 63–69.
Lee J.I., Kim Y.J., Bae H., Cho S.S., Lee J.-H., Kwon S.-T. 2010. Biochemical properties and PCR performance of a family B DNA polymerase from hyperthermophilic euryarchaeon Thermococcus peptonophilus. Appl. Biochem. Biotechnol. 160, 1585–1599.
Harrell R.A., Hart R.P. 1994. Rapid preparation of Thermus flavus DNA polymerase. Genome Res. 3, 372–375.
Carballeira N., Nazabal M., Brito J., Garcia O. 1990. Purification of a thermostable DNA polymerase from Thermus thermophilus HB8, useful in the polymerase chain reaction. Biotechniques. 9, 276–281.
Yang S.-W., Astatke M., Potter J., Chatterjee D.K. 2002. Mutant Thermotoga neapolitana DNA polymerase I: altered catalytic properties for non-templated nucleotide addition and incorporation of correct nucleotides. Nucleic Acids Res. 30, 4314–4320.
Steitz T.A. 1993. DNA- and RNA-dependent DNA polymerases. Curr. Opin. Struct. Biol. 3, 31–38.
Steitz T.A. 1998. A mechanism for all polymerases. Nature. 391, 231–2323.
Steitz T.A. 1999. DNA polymerases: structural diversity and common mechanisms. J. Biol. Chem. 274, 17395–17398.
Joyce C.M. 2013. DNA polymerase I, Bacterial. In Encyclopedia of Biological Chemistry, 2nd ed. Elsevier, pp. 87–90.
Betz K., Malyshev D.A., Lavergne T., Welte W., Diederichs K., Dwyer T.J., Ordoukhanian P., Romesberg F.E., Marx A. 2012. KlenTaq polymerase replicates unnatural base pairs by inducing a Watson–Crick geometry. Nat. Chem. Biol. 8, 612–614.
Raper A.T., Reed A.J., Suo Z. 2018. Kinetic mechanism of DNA polymerases: contributions of conformational dynamics and a third divalent metal ion. Chem. Rev. 118, 6000–6025.
Berdis A.J. 2009. Mechanisms of DNA polymerases. Chem. Rev. 109, 2862–2879.
Brautigam C.A., Steitz T.A. 1998. Structural and functional insights provided by crystal structures of DNA polymerases and their substrate complexes. Curr. Biol. 8, 54–63.
Ignatov K.B., Bashirova A.A., Miroshnikov A.I., Kramarov V.M. 1999. Mutation S543N in the thumb subdomain of the Taq DNA polymerase large fragment suppresses pausing associated with the template structure. FEBS Lett. 448, 145–148.
Drum M., Kranaster R., Ewald C., Blasczyk R., Marx A. 2014. Variants of a Thermus aquaticus DNA polymerase with increased selectivity for applications in allele- and methylation-specific amplification. PLoS One. 9, e96640.
Raghunathan G., Marx A. 2019. Identification of Thermus aquaticus DNA polymerase variants with increased mismatch discrimination and reverse transcriptase activity from a smart enzyme mutant library. Sci. Rep. 9, 590.
Yamagami T., Ishino S., Kawarabayasi Y., Ishino Y. 2014. Mutant Taq DNA polymerases with improved elongation ability as a useful reagent for genetic engineering. Front. Microbiol. 5. 461.
Minnick D.T., Bebenek K., Osheroff W.P., Turner R.M., Astatke M., Liu L., Kunkel T.A., Joyce C.M. 1999. Side chains that influence fidelity at the polymerase active site of Escherichia coli DNA polymerase I (Klenow fragment). J. Biol. Chem. 274, 3067–3075.
Yamagami T., Matsukawa H., Tsunekawa S., Kawarabayasi Y., Ishino S., Ishino Y. 2016. A longer finger-subdomain of family A DNA polymerases found by metagenomic analysis strengthens DNA binding and primer extension abilities. Gene. 576, 690–695.
Roberts R.J. 1995. On base flipping. Cell. 82, 9–12.
Suzuki M., Yoshida S., Adman E.T., Blank A., Loeb L.A. 2000. Thermus aquaticus DNA polymerase I mutants with altered fidelity. J. Biol. Chem. 275, 32728–32735.
Bernad A., Blanco L., Lázaro J., Martín G., Salas M. 1989. A conserved 3′→5′ exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell. 59, 219–228.
Park Y., Choi H., Lee D.S., Kim Y. 1997. Improvement of the 3′–5′ exonuclease activity of Taq DNA polymerase by protein engineering in the active site. Mol. Cells. 7, 419–424.
Ignatov K., Kramarov V., Billingham S. 2009. Chimeric DNA polymerase. US Patent US20090209005A1.
Stenesh J., McGowan G.R. 1977. DNA polymerase from mesophilic and thermophilic bacteria. Biochim. Biophys. Acta, Nucleic Acids Protein Synth. 475, 32–41.
Kevbrin V.V., Zengler K., Lysenko A., Wiegel J. 2005. Anoxybacillus kamchatkensis sp. nov., a novel thermophilic facultative aerobic bacterium with a broad pH optimum from the Geyser valley, Kamchatka. Extremophiles. 9, 391–398.
Namsaraev Z., Babasanova O., Dunaevsky Y., Akimov V., Barkhutova D., Gorlenko V.M., Namsaraev B. 2010. Anoxybacillus mongoliensis sp. nov., a novel thermophilic proteinase producing bacterium isolated from alkaline hot spring, central Mongolia. Microbiology (Moscow). 79, 491‒499. https://doi.org/10.1134/S0026261710040119
Funding
The work was supported by the Ministry of Science and Higher Education, agreement no. 075-15-2021-1085.
Author information
Authors and Affiliations
Contributions
A. A. Bulygin and A. A. Kuznetsova contributed equally to this review.
Corresponding author
Ethics declarations
The authors declare no conflict of interest.
This article does not contain any studies involving humans or animals as research subjects.
Rights and permissions
About this article
Cite this article
Bulygin, A.A., Kuznetsova, A.A., Fedorova, O.S. et al. Comparative Analysis of Family A DNA-Polymerases as a Searching Tool for Enzymes with New Properties. Mol Biol 57, 182–192 (2023). https://doi.org/10.1134/S0026893323020048
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893323020048