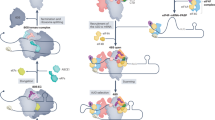
Abstract
The review discusses the role that proteins interacting with the translation termination factors eRF1 and eRF3 play in the control of protein synthesis and prionization. These proteins interact not only with each other, but also with many other proteins involved in controlling the efficiency of translation termination, and associate translation termination with other cell processes. The termination of translation is directly related not only to translation re-initiation and ribosome recycling, but also to mRNA stability and protein quality control. This connection is ensured by the interaction of eRF1 and eRF3 with proteins participating in various cell metabolic processes, such as mRNA transport from the nucleus into the cytoplasm (Dbp5/DDX19 and Gle1), ribosome recycling (Rli1/ABCE1), mRNA degradation (Upf proteins), and translation initiation (Pab1/PABP). In addition to genetic control, there is epigenetic control of translation termination. This mechanism is associated with prion polymerization of the Sup35 protein to form the [PSI+] prion. The maintenance of the [PSI+] prion, like other yeast prions, requires the operation of a system of molecular chaperones and protein sorting factors. The review considers in detail the interaction of the translation termination factors with proteins involved in various cellular processes.




Similar content being viewed by others
REFERENCES
Inge-Vechtomov S., Zhouravleva G., Philippe M. 2003. Eukaryotic release factors (eRFs) history. Biol. Cell. 95, 195–209.
Patino M.M., Liu J.J., Glover J.R., Lindquist S. 1996. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science. 273, 622–626.
Paushkin S.V., Kushnirov V.V., Smirnov V.N., Ter-Avanesyan M.D. 1996. Propagation of the yeast prion-like [PSI +] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 15, 3127–3134.
Glover J.R., Kowal A.S., Schirmer E.C., Patino M.M., Liu J.J., Lindquist S. 1997. Self-seeded fibers formed by Sup35, the protein determinant of [PSI +], a heritable prion-like factor of S. cerevisiae. Cell. 89, 811–819.
King C.Y., Tittmann P., Gross H., Gebert R., Aebi M., Wüthrich K. 1997. Prion-inducing domain 2-114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc. Natl. Acad. Sci. U. S. A. 94, 6618–6622.
Liebman S.W., Chernoff Y.O. 2012. Prions in yeast. Genetics. 191, 1041–1072.
Stansfield I., Jones K.M., Kushnirov V.V., Dagkesamanskaya A.R., Poznyakovski A.I., Paushkin S.V., Nierras C.R., Cox B.S., Ter-Avanesyan M.D., Tuite M.F. 1995. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 14, 4365–4373.
Zhouravleva G., Frolova L., Le Goff X., Le Guellec R., Inge-Vechtomov S., Kisselev L., Philippe M. 1995. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 14, 4065–4072.
Doma M.K., Parker R. 2006. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 440, 561–564.
Cheng Z., Saito K., Pisarev A.V., Wada M., Pisareva V.P., Pestova T.V., Gajda M., Round A., Kong C., Lim M., Nakamura Y., Svergun D.I., Ito K., Song H. 2009. Structural insights into eRF3 and stop codon recognition by eRF1. Genes Dev. 23, 1106–18.
Chen L., Muhlrad D., Hauryliuk V., Cheng Z., Lim M.K., Shyp V., Parker R., Song H. 2010. Structure of the Dom34-Hbs1 complex and implications for no-go decay. Nat. Struct. Mol. Biol. 17, 1233–1240.
Becker T., Armache J.-P., Jarasch A., Anger A.M., Villa E., Sieber H., Motaal B.A., Mielke T., Berninghausen O., Beckmann R. 2011. Structure of the no-go mRNA decay complex Dom34–Hbs1 bound to a stalled 80S ribosome. Nat. Struct. Mol. Biol. 18, 715–720.
Frolova L., Le Goff X., Rasmussen H.H., Cheperegin S., Drugeon G., Kress M., Arman I., Haenni A.L., Celis J.E., Philippe M. 1994. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature. 372, 701–703.
Song H., Mugnier P., Das A.K., Webb H.M., Evans D.R., Tuite M.F., Hemmings B.A., Barford D. 2000. The crystal structure of human eukaryotic release factor eRF1-mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 100, 311–321.
Ito K. 1996. Conserved motifs in prokaryotic and eukaryotic polypeptide release factors: tRNA-protein mimicry hypothesis. Proc. Natl. Acad. Sci. U. S. A. 93, 5443–5448.
Hellen C.U.T. 2018. Translation termination and ribosome recycling in eukaryotes. Cold Spring Harb. Perspect. Biol. 10, a032656.
Mantsyzov A.B., Ivanova E.V., Birdsall B., Kolosov P.M., Kisselev L.L., Polshakov V.I. 2007. NMR assignments of the C-terminal domain of human polypeptide release factor eRF1. Biomol. NMR Assign. 1, 183–185.
Frolova L.Y., Tsivkovskii R.Y., Sivolobova G.F., Oparina N.Y., Serpinsky O.I., Blinov V.M., Tatkov S.I., Kisselev L.L. 1999. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA. 5, 1014–1020.
Ito K., Ebihara K., Nakamura Y. 1998. The stretch of C-terminal acidic amino acids of translational release factor eRF1 is a primary binding site for eRF3 of fission yeast. RNA. 4, 958–972.
Merkulova T.I., Frolova L.Y., Lazar M., Camonis J., Kisselev L.L. 1999. C-terminal domains of human translation termination factors eRF1 and eRF3 mediate their in vivo interaction. FEBS Lett. 443, 41–47.
Moskalenko S.E., Chabelskaya S.V., Inge-Vechtomov S.G., Philippe M., Zhouravleva G.A. 2003. Viable nonsense mutants for the essential gene SUP45 of Saccharomyces cerevisiae. BMC Mol. Biol. 4, 2.
Eurwilaichitr L., Graves F.M., Stansfield I., Tuite M.F. 1999. The C-terminus of eRF1 defines a functionally important domain for translation termination in Saccharomyces cerevisiae. Mol. Microbiol. 32, 485–496.
Andjelković N., Zolnierowicz S., Van Hoof C., Goris J., Hemmings B.A. 1996. The catalytic subunit of protein phosphatase 2A associates with the translation termination factor eRF1. EMBO J. 15, 7156–7167.
Kononenko A.V., Mitkevich V.A., Dubovaya V.I., Kolosov P.M., Makarov A.A., Kisselev L.L. 2008. Role of the individual domains of translation termination factor eRF1 in GTP binding to eRF3. Proteins. 70, 388–393.
Mitkevich V.A., Kononenko A.V., Petrushanko I.Y., Yanvarev D.V., Makarov A.A., Kisselev L.L. 2006. Termination of translation in eukaryotes is mediated by the quaternary eRF1*eRF3*GTP*Mg2+ complex. The biological roles of eRF3 and prokaryotic RF3 are profoundly distinct. Nucleic Acids Res. 34, 3947–3954.
Alkalaeva E.Z., Pisarev A.V., Frolova L.Y., Kisselev L.L., Pestova T.V. 2006. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 125, 1125–1136.
Hauryliuk V., Zavialov A., Kisselev L., Ehrenberg M. 2006. Class-1 release factor eRF1 promotes GTP binding by class-2 release factor eRF3. Biochimie. 88, 747–757.
Frolova L., Le Goff X., Zhouravleva G., Davydova E., Philippe M., Kisselev L. 1996. Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA. 2, 334–341.
Pisareva V.P., Pisarev A.V., Hellen C.U.T., Rodnina M.V., Pestova T.V. 2006. Kinetic analysis of interaction of eukaryotic release factor 3 with guanine nucleotides. J. Biol. Chem. 281, 40224–40235.
Hoshino S., Imai M., Mizutani M., Kikuchi Y., Hanaoka F., Ui M., Katada T. 1998. Molecular cloning of a novel member of the eukaryotic polypeptide chain-releasing factors (eRF). J. Biol. Chem. 273, 22254–22259.
Zhouravleva G., Schepachev V., Petrova A., Tarasov O., Inge-Vechtomov S. 2006. Evolution of translation termination factor eRF3: is GSPT2 generated by retrotransposition of GSPT1’s mRNA? IUBMB Life. 58, 199–202.
Kushnirov V.V., Ter-Avanesyan M.D., Telckov M.V., Surguchov A.P., Smirnov V.N., Inge-Vechtomov S.G. 1988. Nucleotide sequence of the SUP2 (SUP35) gene of Saccharomyces cerevisiae. Gene. 66, 45–54.
Cosson B., Couturier A., Chabelskaya S., Kiktev D., Inge-Vechtomov S., Philippe M., Zhouravleva G. 2002. Poly(A)-binding protein acts in translation termination via eukaryotic release factor 3 interaction and does not influence [PSI +] propagation. Mol. Cell. Biol. 22, 3301–3315.
Urakov V.N., Mitkevich O.V., Safenkova I.V., Ter-Avanesyan M.D. 2017. Ribosome-bound Pub1 modulates stop codon decoding during translation termination in yeast. FEBS J. 284, 1914–1930.
Paushkin S.V., Kushnirov V.V., Smirnov V.N., Ter-Avanesyan M.D. 1997. Interaction between yeast Sup45p (eRF1) and Sup35p (eRF3) polypeptide chain release factors: Implications for prion-dependent regulation. Mol. Cell. Biol. 17, 2798–2805.
Kong C., Ito K., Walsh M.A., Wada M., Liu Y., Kumar S., Barford D., Nakamura Y., Song H. 2004. Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe. Mol. Cell. 14, 233–234.
des Georges A., Hashem Y., Unbehaun A., Grassucci R.A., Taylor D., Hellen C.U.T., Pestova T.V., Frank J. 2014. Structure of the mammalian ribosomal pre-termination complex associated with eRF1.eRF3.GDPNP. Nucleic Acids Res. 42, 3409–3418.
Taylor D., Unbehaun A., Li W., Das S., Lei J., Liao H.Y., Grassucci R.A., Pestova T.V., Frank J. 2012. Cryo-EM structure of the mammalian eukaryotic release factor eRF1–eRF3-associated termination complex. Proc. Natl. Acad. Sci. U. S. A. 109, 18413–18418.
Culbertson M.R., Underbrink K.M., Fink G.R. 1980. Frameshift suppression Saccharomyces cerevisiae: 2. Genetic properties of group II suppressors. Genetics. 95, 833–853.
Leeds P., Peltz S.W., Jacobson A., Culbertson M.R. 1991. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 5, 2303–2314.
Leeds P., Wood J.M., Lee B.S., Culbertson M.R. 1992. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol. Cell. Biol. 12, 2165–2177.
Pulak R., Anderson P. 1993. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 7, 1885–1897.
Cali B.M., Anderson P. 1998. mRNA surveillance mitigates genetic dominance in Caenorhabditis elegans. Mol. Gen. Genet. 260, 176–184.
Page M.F., Carr B., Anders K.R., Grimson A., Anderson P. 1999. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol. Cell. Biol. 19, 5943–5951.
Aronoff R., Baran R., Hodgkin J. 2001. Molecular identification of smg-4, required for mRNA surveillance in C. elegans. Gene. 268, 153–164.
Kashima I., Yamashita A., Izumi N., Kataoka N., Morishita R., Hoshino S., Ohno M., Dreyfuss G., Ohno S. 2006. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 20, 355–367.
Lloyd J.P.B., Davies B. 2013. SMG1 is an ancient nonsense-mediated mRNA decay effector. Plant J. 76, 800–810.
Causier B., Li Z., De Smet R., Lloyd J.P.B., Van de Peer Y., Davies B. 2017. Conservation of nonsense-mediated mRNA decay complex components throughout eukaryotic evolution. Sci. Rep. 7, 16692.
Lloyd J.P.B. 2018. The evolution and diversity of the nonsense-mediated mRNA decay pathway. F1000Research. 7, 1299.
Culbertson M.R., Leeds P.F. 2003. Looking at mRNA decay pathways through the window of molecular evolution. Curr. Opin. Genet. Dev. 13, 207–214.
Atkin A.L., Schenkman L.R., Eastham M., Dahlseid J.N., Lelivelt M.J., Culbertson M.R. 1997. Relationship between yeast polyribosomes and Upf proteins required for nonsense mRNA decay. J. Biol. Chem. 272, 22163–22172.
Altamura N., Groudinsky O., Dujardin G., Slonimski P.P. 1992. NAM7 nuclear gene encodes a novel member of a family of helicases with a Zn-ligand motif and is involved in mitochondrial functions in Saccharomyces cerevisiae. J. Mol. Biol. 224, 575–587.
Weng Y., Czaplinski K., Peltz S.W. 1996. Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol. Cell. Biol. 16, 5477–5490.
Johansson M.J.O., Jacobson A. 2010. Nonsense-mediated mRNA decay maintains translational fidelity by limiting magnesium uptake. Genes Dev. 24, 1491–1495.
He F., Jacobson A. 1995. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 9, 437–454.
He F., Brown A.H., Jacobson A. 1997. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol. Cell. Biol. 17, 1580–1594.
Weng Y., Czaplinski K., Peltz S.W. 1996. Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf protein complex but not mRNA turnover. Mol. Cell. Biol. 16, 5491–5506.
Czaplinski K., Ruiz-Echevarria M.J., Paushkin S. V, Han X., Weng Y., Perlick H.A., Dietz H.C., Ter-Avanesyan M.D., Peltz S.W. 1998. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 12, 1665–1677.
Ivanov P.V., Gehring N.H., Kunz J.B., Hentze M.W., Kulozik A.E. 2008. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 27, 736–747.
Zhouravleva G.A., Gryzina V.A. 2012. Influence of UPF genes on severity of SUP45 mutations.Mol. Biol. (Moscow). 46 (2), 258–269.
Wang W., Czaplinski K., Rao Y., Peltz S.W. 2001. The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J. 20, 880–890.
Serdar L.D., Whiteside D.L., Baker K.E. 2016. ATP hydrolysis by UPF1 is required for efficient translation termination at premature stop codons. Nat. Commun. 7, 14021.
Schuller A.P., Zinshteyn B., Enam S.U., Green R. 2018. Directed hydroxyl radical probing reveals Upf1 binding to the 80S ribosomal E site rRNA at the L1 stalk. Nucleic Acids Res. 46, 2060–2073.
Neu-Yilik G., Raimondeau E., Eliseev B., Yeramala L., Amthor B., Deniaud A., Huard K., Kerschgens K., Hentze M.W., Schaffitzel C., Kulozik A.E. 2017. Dual function of UPF3B in early and late translation termination. EMBO J. 36, 2968–2986.
Kadlec J., Guilligay D., Ravelli R.B., Cusack S. 2006. Crystal structure of the UPF2-interacting domain of nonsense-mediated mRNA decay factor UPF1. RNA. 12, 1817–1824.
Kim Y.K., Maquat L.E. 2019. UPFront and center in RNA decay: UPF1 in nonsense-mediated mRNA decay and beyond. RNA. 25, 407–422.
Min E.E., Roy B., Amrani N., He F., Jacobson A. 2013. Yeast Upf1 CH domain interacts with Rps26 of the 40S ribosomal subunit. RNA. 19, 1105–1115.
Cui Y., Hagan K.W., Zhang S., Peltz S.W. 1995. Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes Dev. 9, 423–436.
Mendell J.T., Medghalchi S.M., Lake R.G., Noensie E.N., Dietz H.C. 2000. Novel Upf2p orthologues suggest a functional link between translation initiation and nonsense surveillance complexes. Mol. Cell. Biol. 20, 8944–8957.
Fourati Z., Roy B., Millan C., Coureux P.-D., Kervestin S., van Tilbeurgh H., He F., Usón I., Jacobson A., Graille M. 2014. A highly conserved region essential for NMD in the Upf2 N-terminal domain. J. Mol. Biol. 426, 3689–3702.
He F., Brown A.H., Jacobson A. 1996. Interaction between Nmd2p and Upf1p is required for activity but not for dominant-negative inhibition of the nonsense-mediated mRNA decay pathway in yeast. RNA. 2, 153–170.
López-Perrote A., Castaño R., Melero R., Zamarro T., Kurosawa H., Ohnishi T., Uchiyama A., Aoyagi K., Buchwald G., Kataoka N., Yamashita A., Llorca O. 2016. Human nonsense-mediated mRNA decay factor UPF2 interacts directly with eRF3 and the SURF complex. Nucleic Acids Res. 44, 1909–1923.
Lykke-Andersen J., Shu M.D., Steitz J.A. 2000. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 103, 1121–1131.
Shirley R.L., Lelivelt M.J., Schenkman L.R., Dahlseid J.N., Culbertson M.R. 1998. A factor required for nonsense-mediated mRNA decay in yeast is exported from the nucleus to the cytoplasm by a nuclear export signal sequence. J. Cell Sci. 111, 3129–3143.
Shirley R.L., Ford A.S., Richards M.R., Albertini M., Culbertson M.R. 2002. Nuclear import of Upf3p is mediated by importin-alpha/-beta and export to the cytoplasm is required for a functional nonsense-mediated mRNA decay pathway in yeast. Genetics. 161, 1465–1482.
Chabelskaya S., Kiktev D., Inge-Vechtomov S., Philippe M., Zhouravleva G. 2004. Nonsense mutations in the essential gene SUP35 of Saccharomyces cerevisiae are non-lethal. Mol. Genet. Genomics. 272, 297–307.
Chabelskaya S., Gryzina V., Moskalenko S., Le Goff C., Zhouravleva G. 2007. Inactivation of NMD increases viability of sup45 nonsense mutants in Saccharomyces cerevisiae. BMC Mol. Biol. 8, 71.
Chabelskaya S.V., Zhouravleva G.A. 2010. Mutations in the SUP35 gene impair nonsense-mediated mRNA decay. Mol Biol (Moscow). 44 (1), 45–53.
Chamieh H., Ballut L., Bonneau F., Le Hir H. 2008. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat. Struct. Mol. Biol. 15, 85–93.
Blobel G. 1973. A protein of molecular weight 78,000 bound to the polyadenylate region of eukaryotic messenger RNAs. Proc. Natl. Acad. Sci. U. S. A. 70, 924–928.
Adam S.A., Nakagawa T., Swanson M.S., Woodruff T.K., Dreyfuss G. 1986. mRNA polyadenylate-binding protein: gene isolation and sequencing and identification of a ribonucleoprotein consensus sequence. Mol. Cell. Biol. 6, 2932–2943.
Sachs A.B., Bond M.W., Kornberg R.D. 1986. A single gene from yeast for both nuclear and cytoplasmic polyadenylate-binding proteins: Domain structure and expression. Cell. 45, 827–835.
Mangus D.A., Evans M.C., Jacobson A. 2003. Poly(A)-binding proteins: Multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 4, 223.
Kühn U., Wahle E. 2004. Structure and function of poly(A)-binding proteins. Biochim. Biophys. Acta. 1678, 67–84.
Sachs A.B., Davis R.W., Kornberg R.D. 1987. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol. Cell. Biol. 7, 3268–3276.
Kozlov G., Siddiqui N., Coillet-Matillon S., Trempe J.-F., Ekiel I., Sprules T., Gehring K. 2002. Solution structure of the orphan PABC domain from Saccharomyces cerevisiae poly(A)-binding protein. J. Biol. Chem. 277, 22822–22828.
Tarun S.Z., Sachs A.B. 1996. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 15, 7168–7177.
Wells S.E., Hillner P.E., Vale R.D., Sachs A.B. 1998. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell. 2, 135–140.
Sachs A.B., Davis R.W. 1989. The poly(A)-binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 58, 857–867.
Munroe D., Jacobson A. 1990. mRNA poly(A) tail, a 3′-enhancer of translational initiation. Mol. Cell. Biol. 10, 3441–3455.
Gallie D.R. 1991. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 5, 2108–2116.
Tarun S.Z., Wells S.E., Deardorff J.A., Sachs A.B. 1997. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc. Natl. Acad. Sci. U. S. A. 94, 9046–9051.
Imataka H., Gradi A., Sonenberg N. 1998. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 17, 7480–7489.
Kessler S.H., Sachs A.B. 1998. RNA recognition motif 2 of yeast Pab1p is required for its functional interaction with eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 18, 51–57.
Otero L.J., Ashe M.P., Sachs A.B. 1999. The yeast poly(A)-binding protein Pab1p stimulates in vitro poly(A)-dependent and cap-dependent translation by distinct mechanisms. EMBO J. 18, 3153–3163.
Richardson R., Denis C.L., Zhang C., Nielsen M.E.O., Chiang Y.-C., Kierkegaard M., Wang X., Lee D.J., Andersen J.S., Yao G. 2012. Mass spectrometric identification of proteins that interact through specific domains of the poly(A)-binding protein. Mol. Genet. Genomics. 287, 711–730.
Hoshino S., Imai M., Kobayashi T., Uchida N., Katada T. 1999. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3'-Poly(A) tail of mRNA. Direct association of erf3/GSPT with polyadenylate-binding protein. J. Biol. Chem. 274, 16677–16680.
Cosson B., Couturier A., Le Guellec R., Moreau J., Chabelskaya S., Zhouravleva G., Philippe M. 2002. Characterization of the poly(A) binding proteins expressed during oogenesis and early development of Xenopus laevis. Biol. Cell. 94, 217–231.
Cosson B., Berkova N., Couturier A., Chabelskaya S., Philippe M., Zhouravleva G. 2002. Poly(A)-binding protein and eRF3 are associated in vivo in human and Xenopus cells. Biol. Cell. 94, 205–216.
Uchida N., Hoshino S.-I., Imataka H., Sonenberg N., Katada T. 2002. A novel role of the mammalian GSPT/eRF3 associating with poly(A)-binding protein in cap/poly(A)-dependent translation. J. Biol. Chem. 277, 50286–50292.
Ivanov A., Mikhailova T., Eliseev B., Yeramala L., Sokolova E., Susorov D., Shuvalov A., Schaffitzel C., Alkalaeva. E. 2016. PABP enhances release factor recruitment and stop codon recognition during translation termination. Nucleic Acids Res. 44, 7766–7776.
Albrecht M., Lengauer T. 2004. Survey on the PABC recognition motif PAM2. Biochem. Biophys. Res. Commun. 316, 129–138.
Brito M., Malta-Vacas J., Carmona B., Aires C., Costa P., Martins A.P., Ramos S., Conde A.R., Monteiro C. 2005. Polyglycine expansions in eRF3/GSPT1 are associated with gastric cancer susceptibility. Carcinogenesis. 26, 2046–2049.
Malta-Vacas J., Chauvin C., Gonçalves L., Nazaré A., Carvalho C., Monteiro C., Bagrel D., Jean-Jean O., Brito M. 2009. eRF3a/GSPT1 12-GGC allele increases the susceptibility for breast cancer development. Oncol. Rep. 21, 1551–1558.
Miri M., Hemati S., Safari F., Tavassoli M. 2012. GGCn polymorphism of eRF3a/GSPT1 gene and breast cancer susceptibility. Med. Oncol. 29, 1581–1585.
Jerbi S., Jolles B., Bouceba T., Jean-Jean O. 2016. Studies on human eRF3–PABP interaction reveal the influence of eRF3a N-terminal glycin repeat on eRF3-PABP binding affinity and the lower affinity of eRF3a 12-GGC allele involved in cancer susceptibility. RNA Biol. 13, 306–315.
Kozlov G., Trempe J.F., Khaleghpour K., Kahvejian A., Ekiel I., Gehring K. 2001. Structure and function of the C-terminal PABC domain of human poly(A)-binding protein. Proc. Natl. Acad. Sci. U. S. A. 98, 4409–4413.
Xie J., Kozlov G., Gehring K. 2014. The “tale” of poly(A) binding protein: The MLLE domain and PAM2-containing proteins. Biochim. Biophys. Acta. 1839, 1062–1068.
Kozlov G., De Crescenzo G., Lim N.S., Siddiqui N., Fantus D., Kahvejian A., Trempe J-F., Elias D., Ekiel I., Sonenberg N., O’Connor-McCourt M., Gehring K. 2004. Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase. EMBO J. 23, 272–281.
Kononenko A.V., Mitkevich V.A., Atkinson G.C., Tenson T., Dubovaya V.I., Frolova L.Y., Makarov A.A., Hauryliuk V. 2010. GTP-dependent structural rearrangement of the eRF1:eRF3 complex and eRF3 sequence motifs essential for PABP binding. Nucleic Acids Res. 38, 548–558.
Kozlov G., Gehring K. 2010. Molecular basis of eRF3 recognition by the MLLE domain of poly(A)-binding protein. PLoS One. 5, e10169.
Hegde R., Srinivasula S.M., Datta P., Madesh M., Wassell R., Zhang Z., Cheong N., Nejmeh J., Fernandes-Alnemri T.,Hoshino S., Alnemri E.S. 2003. The polypeptide chain-releasing factor GSPT1/eRF3 is proteolytically processed into an IAP-binding protein. J. Biol. Chem. 278, 38699–38706.
Hosoda N., Kobayashi T., Uchida N., Funakoshi Y., Kikuchi Y., Hoshino S., Katada T. 2003. Translation termination factor eRF3 mediates mRNA decay through the regulation of deadenylation. J. Biol. Chem. 278, 38287–38291.
Funakoshi Y., Doi Y., Hosoda N., Uchida N., Osawa M., Shimada I., Tsujimoto M., Suzuki T., Katada T., Hoshino S. 2007. Mechanism of mRNA deadenylation: Evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. Genes Dev. 21, 3135–3148.
Osawa M., Hosoda N., Nakanishi T., Uchida N., Kimura T., Imai S., Machiyama A., Katada T., Hoshino S., Shimada I. 2012. Biological role of the two overlapping poly(A)-binding protein interacting motifs 2 (PAM2) of eukaryotic releasing factor eRF3 in mRNA decay. RNA. 18, 1957–1967.
Mangus D.A., Evans M.C., Agrin N.S., Smith M., Gongidi P., Jacobson A. 2004. Positive and negative regulation of poly(A) nuclease. Mol. Cell. Biol. 24, 5521–5533.
Ivanov A., Shuvalova E., Egorova T., Shuvalov A., Sokolova E., Bizyaev N., Shatsky I., Terenin I., Alkalaeva E. 2019. Polyadenylate–binding protein–interacting proteins PAIP1 and PAIP2 affect translation termination. J. Biol. Chem. 294, 8630–8639.
Geourjon C., Deléage G. 1994. SOPM: A self-optimized method for protein secondary structure prediction. Protein Eng. 7, 157–164.
Pisarev A.V., Skabkin M.A., Pisareva V.P., Skabkina O.V., Rakotondrafara A.M., Hentze M.W., Hellen C.U.T., Pestova T.V. 2010. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol. Cell. 37, 196–210.
Shoemaker C.J., Green R. 2011. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc. Natl. Acad. Sci. U. S. A. 108, E1392–E1398.
Barthelme D., Scheele U., Dinkelaker S., Janoschka A., Macmillan F., Albers S.-V., Driessen A.J.M., Stagni M.S., Bill E., Meyer-Klaucke W., Schünemann V., Tampé R. 2007. Structural organization of essential iron-sulfur clusters in the evolutionarily highly conserved ATP-binding cassette protein ABCE1. J. Biol. Chem. 282, 14598–14607.
Khoshnevis S., Gross T., Rotte C., Baierlein C., Ficner R., Krebber H. 2010. The iron-sulphur protein RNase L inhibitor functions in translation termination. EMBO Rep. 11, 214–219.
Becker T., Franckenberg S., Wickles S., Shoemaker C.J., Anger A.M., Armache J.-P., Sieber H., Ungewickell C., Berninghausen O., Daberkow I., Karcher A., Thomm M., Hopfner K-P., Green R., Beckmann R. 2012. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature. 482, 501–506.
Snay-Hodge C.A., Colot H.V., Goldstein A.L., Cole C.N. 1998. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J. 17, 2663–2676.
Tseng S.S., Weaver P.L., Liu Y., Hitomi M., Tartakoff A.M., Chang T.H. 1998. Dbp5p, a cytosolic RNA helicase, is required for poly(A)+ RNA export. EMBO J. 17, 2651–2662.
Tieg B., Krebber H. 2013. Dbp5—from nuclear export to translation. Biochim. Biophys. Acta. 1829, 791–798.
Estruch F., Hodge C., Gómez-Navarro N., Peiró-Chova L., Heath C.V., Cole C.N. 2012. Insights into mRNP biogenesis provided by new genetic interactions among export and transcription factors. BMC Genet. 13, 80.
Schmitt C., von Kobbe C., Bachi A., Panté N., Rodrigues J.P., Boscheron C., Rigaut G., Wilm M., Séraphin B., Carmo-Fonseca M., Izaurralde E. 1999. Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. EMBO J. 18, 4332–4347.
Gross T., Siepmann A., Sturm D., Windgassen M., Scarcelli J.J., Seedorf M., Cole C.N., Krebber H. 2007. The DEAD-box RNA helicase Dbp5 functions in translation termination. Science. 315, 646–649.
Murphy R., Wente S.R. 1996. An RNA-export mediator with an essential nuclear export signal. Nature. 383, 357–360.
Watkins J.L., Murphy R., Emtage J.L., Wente S.R. 1998. The human homologue of Saccharomyces cerevisiae Gle1p is required for poly(A)+ RNA export. Proc. Natl. Acad. Sci. U. S. A. 95, 6779–6784.
Alcázar-Román A.R., Tran E.J., Guo S., Wente S.R. 2006. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat. Cell Biol. 8, 711–716.
Adams R.L., Mason A.C., Glass L., Aditi, Wente S.R. 2017. Nup42 and IP6 coordinate Gle1 stimulation of Dbp5/DDX19B for mRNA export in yeast and human cells. Traffic. 18, 776–790.
Montpetit B., Thomsen N.D., Helmke K.J., Seeliger M.A., Berger J.M., Weis K. 2011. A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature. 472, 238–242.
Bolger T.A., Folkmann A.W., Tran E.J., Wente S.R. 2008. The mRNA export factor Gle1 and inositol hexakisphosphate regulate distinct stages of translation. Cell. 134, 624–633.
Hinnebusch A.G. 2006. eIF3: A versatile scaffold for translation initiation complexes. Trends Biochem. Sci. 31, 553–562.
Pisarev A.V., Hellen C.U.T., Pestova T.V. 2007. Recycling of eukaryotic posttermination ribosomal complexes. Cell. 131, 286–299.
Beznosková P., Cuchalová L., Wagner S., Shoemaker C.J., Gunišová S., von der Haar T., Valášek L.S. 2013. Translation initiation factors eIF3 and HCR1 control translation termination and stop codon read-through in yeast cells. PLoS Genet. 9, e1003962.
Kikuchi Y., Shimatake H., Kikuchi A. 1988. A yeast gene required for the G1-to-S transition encodes a protein containing an A-kinase target site and GTPase domain. EMBO J. 7, 1175–1182.
Tikhomirova V.L., Inge-Vechtomov S.G. 1996. Sensitivity of sup35 and sup45 suppressor mutants in Saccharomyces cerevisiae to the anti-microtubule drug benomyl. Curr. Genet. 30, 44–49.
Basu J., Williams B.C., Li Z., Williams E.V., Goldberg M.L. 1998. Depletion of a Drosophila homolog of yeast Sup35p disrupts spindle assembly, chromosome segregation, and cytokinesis during male meiosis. Cell Motil. Cytoskeleton. 39, 286–302.
Borchsenius A.S., Tchourikova A.A., Inge-Vechtomov S.G. 2000. Recessive mutations in SUP35 and SUP45 genes coding for translation release factors affect chromosome stability in Saccharomyces cerevisiae. Curr. Genet. 37, 285–291.
Valouev I.A., Kushnirov V.V., Ter-Avanesyan M.D. 2002. Yeast polypeptide chain release factors eRF1 and eRF3 are involved in cytoskeleton organization and cell cycle regulation. Cell Motil. Cytoskeleton. 52, 161–173.
Stevens R.C., Davis T.N. 1998. Mlc1p is a light chain for the unconventional myosin Myo2p in Saccharomyces cerevisiae. J. Cell Biol. 142, 711–722.
Valouev I.A., Urakov V.N., Kochneva-Pervukhova N.V., Smirnov V.N., Ter-Avanesyan M.D. 2004. Translation termination factors function outside of translation: Yeast eRF1 interacts with myosin light chain, Mlc1p, to effect cytokinesis. Mol. Microbiol. 53, 687–696.
Bailleul P.A., Newnam G.P., Steenbergen J.N., Chernoff Y.O. 1999. Genetic study of interactions between the cytoskeletal assembly protein Sla1 and prion-forming domain of the release factor Sup35 (eRF3) in Saccharomyces cerevisiae. Genetics. 153, 81–94.
Ganusova E.E., Ozolins L.N., Bhagat S., Newnam G.P., Wegrzyn R.D., Sherman M.Y., Chernoff Y.O. 2006. Modulation of prion formation, aggregation, and toxicity by the actin cytoskeleton in yeast. Mol. Cell. Biol. 26, 617–629.
Manogaran A.L., Hong J.Y., Hufana J., Tyedmers J., Lindquist S., Liebman S.W. 2011. Prion formation and polyglutamine aggregation are controlled by two classes of genes. PLoS Genet. 7, e1001386.
Moosavi B., Mousavi B., Yang G.-F. 2016. Actin, membrane trafficking and the control of prion induction, propagation and transmission in yeast. Traffic. 17, 5–20.
Tyedmers J., Madariaga M.L., Lindquist S. 2008. Prion switching in response to environmental stress. PLoS Biol. 6, e294.
Speldewinde S.H., Doronina V.A., Tuite M.F., Grant C.M. 2017. Disrupting the cortical actin cytoskeleton points to two distinct mechanisms of yeast [PSI +] prion formation. PLoS Genet. 13, e1006708.
Chernova T.A., Romanyuk A.V., Karpova T.S., Shanks J.R., Ali M., Moffatt N., Howie R.L., O’Dell A., McNally J.G., Liebman S.W., Chernoff Y.O., Wilkinson K.D. 2011. Prion induction by the short-lived, stress-induced protein Lsb2 is regulated by ubiquitination and association with the actin cytoskeleton. Mol. Cell. 43, 242–252.
Chernova T.A., Kiktev D.A., Romanyuk A.V., Shanks J.R., Laur O., Ali M., Ghosh A., Kim D., Yang Z., Mang M., Chernoff Y.O., Wilkinson K.D. 2017. Yeast short-lived actin-associated protein forms a metastable prion in response to thermal stress. Cell Rep. 18, 751–761.
Ali M., Chernova T.A., Newnam G.P., Yin L., Shanks J., Karpova T.S., Lee A., Laur O., Subramanian S., Kim D., McNally J.G., Seyfried N.T., Chernoff Y.O., Wilkinson K.D. 2014. Stress-dependent proteolytic processing of the actin assembly protein Lsb1 modulates a yeast prion. J. Biol. Chem. 289, 27625–27639.
Li X., Rayman J.B., Kandel E.R., Derkatch I.L. 2014. Functional role of Tia1/Pub1 and Sup35 prion domains: directing protein synthesis machinery to the tubulin cytoskeleton. Mol. Cell. 55, 305–318.
Derkatch I.L., Bradley M.E., Zhou P., Chernoff Y.O., Liebman S.W. 1997. Genetic and environmental factors affecting the de novo appearance of the [PSI +] prion in Saccharomyces cerevisiae. Genetics. 147, 507–519.
Sondheimer N., Lindquist S. 2000. Rnq1: An epigenetic modifier of protein function in yeast. Mol. Cell. 5, 163–172.
Derkatch I.L., Bradley M.E., Hong J.Y., Liebman S.W. 2001. Prions affect the appearance of other prions: The story of [PIN +]. Cell. 106, 171–182.
Derkatch I.L., Bradley M.E., Masse S.V., Zadorsky S.P., Polozkov G.V., Inge-Vechtomov S.G., Liebman S.W. 2000. Dependence and independence of [PSI +] and [PIN +]: A two-prion system in yeast? EMBO J. 19, 1942–1952.
Derkatch I.L., Uptain S.M., Outeiro T.F., Krishnan R., Lindquist S.L., Liebman S.W. 2004. Effects of Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation of the [PSI +] prion in yeast and aggregation of Sup35 in vitro. Proc. Natl. Acad. Sci. U. S. A. 101, 12934–12939.
Bagriantsev S., Liebman S.W. 2004. Specificity of prion assembly in vivo: [PSI +] and [PIN +] form separate structures in yeast. J. Biol. Chem. 279, 51042–51048.
Bondarev S.A., Likholetova D.V., Belousov M.V., Zhouravleva G.A. 2017. Rnq1 protein protects [PSI +] prion from effect of the PNM mutation. Mol. Biol. (Moscow). 51 (2), 323–327.
Osherovich L.Z., Weissman J.S. 2001. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI +] prion. Cell. 106, 183–194.
Wickner R. 1994. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 264, 566–569.
Du Z., Park K.-W., Yu H., Fan Q., Li L. 2008. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat. Genet. 40, 460–465.
Du Z., Li L. 2014. Investigating the interactions of yeast prions: [SWI +], [PSI +], and [PIN +]. Genetics. 197, 685–700.
Patel B.K., Gavin-Smyth J., Liebman S.W. 2009. The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat Cell Biol. 11, 344–349.
Kurahashi H., Oishi K., Nakamura Y. 2011. A bipolar personality of yeast prion proteins. Prion. 5, 305–310.
Yang Z., Hong J.Y., Derkatch I.L., Liebman S.W. 2013. Heterologous Gln/Asn-rich proteins impede the propagation of yeast prions by altering chaperone availability. PLoS Genet. 9, e1003236.
Derkatch I.L., Liebman S.W. 2013. The story of stolen chaperones: How overexpression of Q/N proteins cures yeast prions. Prion. 7, 294–300.
Bagriantsev S.N., Gracheva E.O., Richmond J.E., Liebman S.W. 2008. Variant-specific [PSI +] infection is transmitted by Sup35 polymers within [PSI +] aggregates with heterogeneous protein composition. Mol. Biol. Cell. 19, 2433–2443.
Nevzglyadova O.V., Artemov A.V., Mittenberg A.G., Solovyov K.V., Kostyleva E.I., Mikhailova E.V., Kuznetsova I.M., Turoverov K.K., Soidla T.R. 2009. Prion-associated proteins in yeast: Comparative analysis of isogenic [PSI +] and [psi –] strains. Yeast. 26, 611–631.
Chernova T.A., Wilkinson K.D., Chernoff Y.O. 2014. Physiological and environmental control of yeast prions. FEMS Microbiol. Rev. 38, 326–344.
Kushnirov V.V., Ter-Avanesyan M.D. 1998. Structure and replication of yeast prions. Cell. 94, 13–16.
Matveenko A.G., Barbitoff Y.A., Jay-Garcia L.M., Chernoff Y.O., Zhouravleva G.A. 2018. Differential effects of chaperones on yeast prions: CURrent view. Curr. Genet. 64, 317–325.
Chernoff Y., Lindquist S., Ono B., Inge-Vechtomov S., Liebman S. 1995. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [PSI +]. Science. 268, 880–884.
Ferreira P.C., Ness F., Edwards S.R., Cox B.S., Tuite M.F. 2001. The elimination of the yeast [PSI +] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol. Microbiol. 40, 1357–1369.
Higurashi T., Hines J.K., Sahi C., Aron R., Craig E.A. 2008. Specificity of the J-protein Sis1 in the propagation of 3 yeast prions. Proc. Natl. Acad. Sci. U. S. A. 105, 16596–16601.
Kryndushkin D.S., Smirnov V.N., Ter-Avanesyan M.D., Kushnirov V.V. 2002. Increased expression of Hsp40 chaperones, transcriptional factors, and ribosomal protein Rpp0 can cure yeast prions. J. Biol. Chem. 277, 23702–23708.
Barbitoff Y.A., Matveenko A.G., Moskalenko S.E., Zemlyanko O.M., Newnam G.P., Patel A., Chernova T.A., Chernoff Y.O., Zhouravleva G.A. 2017. To CURe or not to CURe? Differential effects of the chaperone sorting factor Cur1 on yeast prions are mediated by the chaperone Sis1. Mol. Microbiol. 105, 242–257.
Kiktev D.A., Patterson J.C., Muller S., Bariar B., Pan T., Chernoff Y.O. 2012. Regulation of chaperone effects on a yeast prion by cochaperone Sgt2. Mol. Cell. Biol. 32, 4960–4970.
Liu B., Larsson L., Caballero A., Hao X., Oling D., Grantham J., Nyström T. 2010. The polarisome is required for segregation and retrograde transport of protein aggregates. Cell. 140, 257–267.
Newnam G.P., Birchmore J.L., Chernoff Y.O. 2011. Destabilization and recovery of a yeast prion after mild heat shock. J. Mol. Biol. 408, 432–448.
Newnam G.P., Wegrzyn R.D., Lindquist S.L., Chernoff Y.O. 1999. Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol. Cell. Biol. 19, 1325–1333.
Chernoff Y.O., Newnam G.P., Kumar J., Allen K., Zink A.D. 1999. Evidence for a protein mutator in yeast: Role of the Hsp70-related chaperone ssb in formation, stability, and toxicity of the [PSI +] prion. Mol. Cell. Biol. 19, 8103–8112.
Allen K.D., Wegrzyn R.D., Chernova T.A., Müller S., Newnam G.P., Winslett P.A., Wittich, K.B., Wilkinson K.D., Chernoff Y.O. 2005. Hsp70 chaperones as modulators of prion life cycle: Novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI +]. Genetics. 169, 1227–1242.
Kiktev D.A., Melomed M.M., Lu C.D., Newnam G.P., Chernoff Y.O. 2015. Feedback control of prion formation and propagation by the ribosome-associated chaperone complex. Mol. Microbiol. 96, 621–632.
Wickner R.B., Edskes H.K., Son M., Wu S., Niznikiewicz M. 2020. How do yeast cells contend with prions? Int. J. Mol. Sci. 21, 4742.
Wickner R.B., Kelly A.C., Bezsonov E.E., Edskes H.K. 2017. [PSI +] prion propagation is controlled by inositol polyphosphates. Proc. Natl. Acad. Sci. U. S. A. 114, E8402–E8410.
Son M., Wickner R.B. 2018. Nonsense-mediated mRNA decay factors cure most [PSI +] prion variants. Proc. Natl. Acad. Sci. U. S. A. 115, E1184–E1193.
Matveenko A.G., Belousov M.V., Bondarev S.A., Moskalenko S.E., Zhouravleva G.A. 2016. Identification of new genes that affect [PSI +] prion toxicity in Saccharomyces cerevisiae yeast. Mol. Biol. (Moscow). 50 (5), 710–718.
Matveenko A.G., Drozdova P.B., Belousov M.V, Moskalenko S.E., Bondarev S.A., Barbitoff Y.A., Nizhnikov A.A., Zhouravleva G.A. 2016. SFP1-mediated prion-dependent lethality is caused by increased Sup35 aggregation and alleviated by Sis1. Genes Cells. 21, 1290–1308.
Urakov V.N., Mitkevich O.V., Dergalev A.A., Ter-Avanesyan M.D. 2018. The Pub1 and Upf1 proteins act in concert to protect yeast from toxicity of the [PSI +]. Int. J. Mol. Sci. 19, 3663.
Sergeeva A.V., Sopova J.V., Belashova T.A., Siniukova V.A., Chirinskaite A.V., Galkin A.P., Zadorsky S.P. 2019. Amyloid properties of the yeast cell wall protein Toh1 and its interaction with prion proteins Rnq1 and Sup35. Prion. 13, 21–32.
Funding
This work was supported by the Russian Foundation for Basic Research (project no. 20-14-50271).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests. The authors declare that they have no conflict of interest.
This work does not contain any studies involving animals or human subjects performed by any of the authors.
Additional information
Translated by T. Tkacheva
Abbreviations: aa, amino acid residues (with digits); RRM, RNA recognition motif; NMD, nonsense-mediated decay; IPOD, insoluble protein deposit.
Rights and permissions
About this article
Cite this article
Zhouravleva, G.A., Bondarev, S.A., Zemlyanko, O.M. et al. Role of Proteins Interacting with the eRF1 and eRF3 Release Factors in the Regulation of Translation and Prionization. Mol Biol 56, 147–165 (2022). https://doi.org/10.1134/S0026893322010101
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893322010101