Abstract
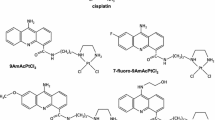
Composite sequence-specific ligands with DNA-damaging groups may dramatically increase the efficacy of radiotherapy. The most promising damage sensitizers are the atoms of heavy elements, in which electrons are emitted from upper orbitals and a multiply charged positive ion forms when an electron is kicked out from lower orbitals. The biophysical mechanisms of DNA damage produced by these sensitizers are far from fully understood. In this work, high-performance polyacrylamide gel electrophoresis (PAGE) in denaturing gel was used to investigate the nature of DNA cleavage on exposure to β radiation for complexes of heavy atom-containing ligands with DNA restriction fragments. It was demonstrated for the first time that DNA in complexes with Pt-bis-netropsin or mercury salts is cleaved in the vicinity of the heavy atom in the presence of radioactive isotopes emitting β particles of different energies. In the presence of 1M glycerol, the cleavage of the DNA sugar-phosphate backbone was almost entirely due to a neutralization of the multiply charged Auger ion and was not associated with the Auger electron electron-beam radiolysis. Based on the observations, a relatively simple technique was proposed for precise localization of binding sites for various DNA ligands containing a heavy atom. Analysis of the end groups at the cleavage point and the nature of damage to the complementary DNA chain made it possible to speculate about the mechanisms of direct influence of irradiation on a heavy atom.












Similar content being viewed by others
REFERENCES
Gursky G.V., Zasedatelev A.S., Zhuze A.L., Khorlin A.A., Grokhovsky S.L., Streltsov S.A., Surovaya A.N., Nikitin S.M., Krylov A.S., Retchinsky V.O., Mikhailov M.V., Beabealashvili R.S., Gottich B.P. 1983. Synthetic sequence-specific ligands. Cold Spring Harb. Symp. Quant. Biol. 47, 367−378.
Zimmer C., Wähnert U. 1986. Nonintercalating DNA-binding ligands: Specificity of the interaction and their use as tools in biophysical, biochemical and biological investigations of the genetic material. Progr. Biophys. Mol. Biol. 47, 31−112.
Surovaya A.N., Grokhovsky S.L., Bazhulina N.P., Gursky G.V. 2008. DNA-binding activity of bis-netropsin containing a cis-diaminoplatinum group between two netropsin fragments. Biophysics (Moscow). 53, 344–351.
Waring M.J., Neidle, S. 2006. Sequence-Specific DNA Binding Agents. RSC Biomolecular Sciences, vol. 6. London: RSC Publishing.
Blackledge M. S., Melander C. 2013. Programmable DNA-binding small molecules. Bioorg. Med. Chem. 21, 6101−6114.
Taniguchi J., Pandian G.N., Hidaka T., Hashiya K., Bando T., Kim K.K., Sugiyama H. 2017. A synthetic DNA-binding inhibitor of SOX2 guides human induced pluripotent stem cells to differentiate into mesoderm. Nucleic Acids Res. 45, 9219−9228.
Kim Y. G., Cha J., Chandrasegaran S. 1996. Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. U. S. A. 93, 1156−1160.
Sanjana N.E., Cong L., Zhou Y., Cunniff M.M., Feng G., Zhang F. 2012. A transcription activator-like effector toolbox for genome engineering. Nat. Protoc. 7, 171−192.
Platzman R.L. 1952. In: Symposium on Radiobiology. Ed. Nickson J.J. New York: Wiley, 95−115.
Feinendegen L.E. 1975. Biological damage from the Auger effect, possible benefits. Radiat. Environ. Biophys. 12, 85−99.
Portugal J., Barceló F. 2016. Noncovalent binding to DNA: Still a target in developing anticancer agents. Curr. Med. Chem. 23, 4108−4134.
Martin R.F., Feinendegen L.E. 2016. The quest to exploit the Auger effect in cancer radiotherapy: A reflective review. Int. J. Rad. Biol. 92, 617−632.
Grokhovsky S. L., Zubarev V. E. 1990. Specific cleavage of double-stranded DNA caused by X-ray ionization of the platinum atom. Dokl. Akad. Nauk SSSR. 313, 1500−1504.
Grokhovsky S.L., Zubarev V.E. 1991. Sequence-specific cleavage of double-stranded DNA caused by X-ray ionization of the platinum atom in the Pt-bis-netropsin‒DNA complex. Nucleic Acids Res. 19, 257−264.
Grokhovsky S.L., Nikolaev V.A., Zubarev V.E., Suro-vaia A.N., Zhuze A.L., Chernov B.K., Sidorova N., Zasedatelev A.S. and Gursky G.V., 1992. Specific DNA cleavage by an analog of netropsin containing a copper(II) chelating peptide Gly-Gly-His. Mol. Biol. (Moscow). 26, 1274−1297.
Nikolaev V.A., Surovaya A.N., Sidorova Grokhovsky S.L., Zasedatelev A.S., Gursky G.V., Zhuze A.L. 1993. DNA-base-pair sequence-specific ligands: 10. Synthesis and binding to DNA of netropsin analogs containing a copper-chelating peptide. Mol. Biol. (Moscow). 27, 117−128.
Grokhovsky S.L., Gottikh B.P., Zhuze A.L. 1992. DNA base pair sequence specific ligands: 9. Synthesis of distamycin A and netropsin analogues containing a sarcolysin residue or a platinum(II) atom. Bioorg. Khim. 18, 313−324.
Sevilla M. D., Becker D., Kumar A., Adhikary A. 2016. Gamma and ion-beam irradiation of DNA: Free radical mechanisms, electron effects, and radiation chemical track structure. Rad. Phys. Chem. 128, 60−74.
Yokoya A., Ito T. 2017. Photon-induced Auger effect in biological systems: A review. Int. J. Rad. Biol. 93, 743−756.
Halpern A., Stocklin G. 1977. Chemical and biological consequences of beta-decay. Rad. Environ. Biophys. 14, 257–274.
Alloni D., Cutaia C., Mariotti L., Friedlandd W., Ottolenghi A. 2014. Modeling dose deposition and DNA damage due to low-energy beta-emitters. Radiat. Res. 182, 322–330.
Grokhovsky S., Il’icheva I., Nechipurenko D., Golovkin M., Taranov G., Panchenko L., Polozov R., Nechipurenko Y. 2012. Quantitative analysis of electrophoresis data: Application to sequence-specific ultrasonic cleavage of DNA. In: Gel Electrophoresis-Principles and Basics. London: InTech, pp. 218–238. doi 10.5772/2205
Grokhovsky S.L., Ilicheva I.A., Nechipurenko D.Yu., Golovkin M.V., Panchenko L.A., Polozov R.V., Nechipurenko Y.D. 2011. Sequence-specific ultrasonic cleavage of DNA. Biophys. J. 100, 117−125.
Grokhovsky S.L., Surovaya A.N., Sidorova N.Ju., Gursky G.V. 1989. Synthesis of nonlinear sequence-specific DNA binding peptide with specificity determinants similar to those of 434 Cro repressor. Mol. Biol. (Moscow). 23, 1558−1580.
Grokhovsky S.L. 2006. Specificity of DNA cleavage by ultrasound. Mol. Biol. (Moscow). 40, 276–283.
Das R., Laederach A., Pearlman S.M., Herschlag D., Altman R.B. 2005. SAFA: Semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA. 11, 344−354.
Nechipurenko Yu.D., Golovkin M.V., Nechipurenko D.Yu., Il’icheva I.N., Panchenko L.A., Polozov R.V., Grokhovsky S.L., Gursky G.V. 2008. Quantitative methods for analysis of DNA cleavage by ultrasound. Mathematics, Computer, Education: Proc. 15th Int. Conf. Izhevsk, vol. 3, pp. 26–35.
Tate W.P., Petersen G.B. 1975. Stability of pyrimidine oligodeoxyribonucleotides released during degradation of deoxyribonucleic acid with formic acid–diphenylamine reagent. Biochem. J. 147, 439–445.
Tullius T.D. 1989. Physical studies of protein-DNA complexes by footprinting. Ann. Rev. Biophys. Biophys. Chem. 18, 213−237.
Balasubramanian B., Pogozelski W.K., Tullius T.D. 1998. DNA strand breaking by the hydroxyl radical is governed by the accessible surface areas of the hydrogen atoms of the DNA backbone. Proc. Natl. Acad. Sci. U. S. A. 95, 9738−9743.
Pogozelski W.K., Tullius T.D. 1998. Oxidative strand scission of nucleic acids: Routes initiated by hydrogen abstraction from the sugar moiety. Chem. Rev. 98, 1089−1108.
Purkayastha S., Milligan J.R., Bernhard W.A. 2005. Correlation of free radical yields with strand break yields produced in plasmid DNA by the direct effect of ionizing radiation. J. Phys. Chem. B. 109, 16 967−16 973.
Chan W., Chen B., Wang L., Taghizadeh K., Demott M.S., Dedon P.C. 2010. Quantification of the 2-deoxyribonolactone and nucleoside 5′-aldehyde products of 2‑deoxyribose oxidation in DNA and cells by isotope-dilution gas chromatography mass spectrometry: Differential effects of γ-radiation and Fe2+–EDTA. J. Am. Chem. Soc. 132, 6145−6153.
Schroeder G.K., Lad C., Wyman P., Williams N.H., Wolfenden R. 2006. The time required for water attack at the phosphorus atom of simple phosphodiesters and of DNA. Proc. Natl. Acad. Sci. U. S. A. 103, 4052–4055.
von Sonntag C. 2006. Free-Radical-Induced DNA Damage and Its Repair. Berlin: Springer.
Franchet-Beuzit J., Spotheim-Maurizot M., Sabattier R., Blazy-Baudras B., Charlier M. 1993. Radiolytic footprinting: Beta rays, gamma photons, and fast neutrons probe DNA–protein interactions. Biochemistry. 32, 2104−2110.
Grokhovsky S.L., Il’icheva I.A., Nechipurenko D.Yu., Panchenko L.A., Polozov R.V., Nechipurenko Yu.D. 2008. Ultrasonic cleavage of DNA: Quantitative analysis of sequence specificity. Biophysics (Moscow), 53, 250–252.
Halpern A. 1982. In: Uses of Synchrotron Radiation in Biology. Ed. Stuhrmann H.B. London: Academic, pp. 255–283.
Halpern A., Stocklin G. 1977. Chemical and biological consequences of beta-decay. Radiat. Environ. Biophys. 14, 257−274.
Martin R.F., Pardee M. 1985. Preparation of carrier free [125I] iodoHoechst 33258. Int. J. Appl. Radiat. Isot. 36, 745−747.
Blagoi Yu.P., Galkin V.L., Gladchenko G.O., Kornilova S.V., Sorokin V.A., Schkorbatov A.G. 1991. Metal Complexes of Nucleic Acids in Solutions. Kiev: Naukova Dumka.
Kharatishvili M.G., Esipova N.G., Zhuze A.L., Grokhovskii S.L., Andronikashvili E.L. 1985. Formation of the left-handed helix during simultaneous treatment of poly[d(GC)] with bis-netropsin and Zn(II) ions. Biophysics (Moscow) 30, 701−703.
Leonarski F., D’Ascenzo L., Auffinger P. 2016. Mg2+ ions: Do they bind to nucleobase nitrogens? Nucleic Acids Res. 45. 987−1004.
Hadjiliadis N.D., Sletten E. (Eds.). 2009. Metal Complex–DNA Interactions. Chichester, UK: Wiley.
Gruenwedel D.W., Cruikshank M.K. 1989. Mercury-induced transitions between right-handed and putative left-handed forms of poly[d(AT)•d(AT)] and poly[d(GC)•d[(GC)]. Nucleic Acids Res. 17, 9075–9086.
Grokhovsky S.L., Il’icheva I.A., Panchenko L.A., Golovkin M.V., Polozov R.V., Nechipurenko D.Y. 2013. Ultrasonic cleavage of DNA in complexes with Ag (I), Cu (II), Hg (II). Biophysics (Moscow). 58, 27−36.
Gruenwedel D.W., Cruikshank M.K. 1990. Mercury-induced DNA polymorphism: Probing the conformation of mercury(II)-DNA via staphylococcal nuclease digestion and circular dichroism measurements. Biochemistry. 29, 2110−2116.
Swasey S.M., Leal L.E., Lopez-Acevedo O., Pavlovich J., Gwinn E.G. 2015. Silver(I) as DNA glue: Ag+-mediated guanine pairing revealed by removing Watson–Crick constraints. Sci. Rep. 5, 10 163.
Dairaku T., Furuita K., Sato H., Šebera J., Nakashima K., Ono A., Sychrovský V., Kojima C., Tanaka Y. 2016. Hg(II)/Ag(I)-mediated base pairs and their NMR spectroscopic studies. Inorg. Chim. Acta. 452, 34−42.
Liu H., Cai C., Haruehanroengra P., Yao Q., Chen Y., Yang C., Luo Q., Wu B., Li J., Ma J. Sheng J. 2017. Flexibility and stabilization of Hg(II)-mediated C:T and T:T base pairs in DNA duplex. Nucleic Acids Res. 45, 2910−2918.
Ding W., Xu M., Zhu H., Liang H. 2013. Mechanism of the hairpin folding transformation of thymine-cytosine-rich oligonucleotides induced by Hg(II) and Ag(I) ions. Eur. Phys. J. E. 36, 1−8.
Stepanenko V.F., Yaskova E.K., Belukha I.G., Petriev V.M., Skvortsov V.G., Kolyzhenkov T.V., Petukhov A.D., Dubov D.V. 2015. The calculation of internal irradiation of nano-, micro- and macro-biostructures with electrons, beta particles and quantum radiation of different energy for the development and analysis of new radiopharmaceuticals in nuclear medicine. Radiatsiya i Risk. 24, 35−60.
Lee B.Q., Kibédi T., Stuchbery A.E., Robertson K.A. 2012. Atomic radiations in the decay of medical radioisotopes: A physics perspective. Comp. Math. Methods Med. 2012, ID 651475.
Itala E., Kooser K., Rachlew E., Levola H., Ha D.T., Kukk E. 2015. Gas-phase study on uridine: Conformation and X-ray photofragmentation. J. Chem. Phys. 142, 194 303.
McBride T.J., Preston B.D., Loeb L.A. 1991. Mutagenic spectrum resulting from DNA damage by oxygen radicals. Biochemistry. 30, 207−213.
Lobachevsky P., Clark G.R., Pytel P.D., Leung B., Skene C., Andrau L., White J.M., Karagiannis T., Cullinane C., Lee B.Q., Stuchbery A. 2016. Strand breakage by decay of DNA-bound 124I provides a basis for combined PET imaging and Auger endoradiotherapy. Int. J. Radiat. Biol. 92, 686–697.
Balagurumoorthy P., Xu X., Wang K., Adelstein S.J., Kassis A.I. 2012. Effect of distance between decaying 125I and DNA on Auger-electron induced double-strand break yield. Int. J. Radiat. Biol. 88, 998–1008.
Jahnke T. 2015. Interatomic and intermolecular Coulombic decay: The coming of age story. J. Phys. B: At. Mol. Opt. Phys. 48, 082001.
ACKNOWLEDGMENTS
This work was supported by the Program of Basic Research at Russian Academies of Sciences from 2013 to 2020 (project no. 01201363818) and the Program for molecular and cell biology of the Presidium of the Russian Academy of Sciences.
Author information
Authors and Affiliations
Corresponding author
Additional information
Translated by T. Tkacheva
Rights and permissions
About this article
Cite this article
Grokhovsky, S.L. Use of β Radiation to Localize the Binding Sites of Mercury Ions and Platinum-Containing Ligand in DNA. Mol Biol 52, 732–748 (2018). https://doi.org/10.1134/S0026893318050072
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893318050072