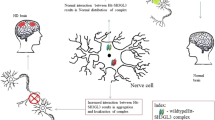
Abstract
Huntingtin (HTT) occurs in the neuronal cytoplasm and can interact with structural elements of synapses. Huntington’s disease (HD) results from pathological expansion of a polyglutamine stretch in the HTT molecule, being probably associated with aberrant protein–protein interactions. The pathogenetic mechanism is still incompletely understood. Alterations of the synaptic structure and plasticity in the hippocampus are observed in early HD. The objective of the study was to theoretically evaluate the HTT contribution to changes in synaptic plasticity by integrating the available experimental data. HTT protein complexes are involved in maintaining the efficiency of synaptic transmission. A pathogenic HTT form (polyQ-HTT) probably disrupts the protein–protein interactions in distorts the dynamics of molecular processes in the synapsis. It was assumed that polyQ-HTT may compete with postsynaptic density proteins and proteins regulating cytoskeleton remodeling.
Similar content being viewed by others
Abbreviations
- LTP:
-
long-term potentiation
- AMPAR:
-
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor
- HD:
-
Huntington’s disease
- CME:
-
clathrin-mediated endocytosis
- HTT:
-
huntingtin
- AP2:
-
adaptor protein 2
- HIP1:
-
huntingtininteracting protein 1
- Pacsin1:
-
protein kinase C and casein kinase substrate in neurons protein 1
- SH3G3:
-
SH3 domaincontaining GRB2-like protein 3
- Dyn:
-
dynamin
- ITSN1:
-
intersectin 1 (EH and SH3 domain protein 1
- ArhGAP31:
-
Rho GTPase-activating protein 31
- N-Was:
-
neural Wiskott–Aldrich syndrome protein
- Hip1R:
-
Huntingtin-interacting protein 1-related protein
- KIF5A:
-
kinesin heavy chain isoform 5A
- Grip1:
-
AMPA receptor subunit–GluR2-interacting protein
- GluR2:
-
GluRS subunit of AMPAR
- HAP1:
-
HTT-associated protein 1
- FRMPD4:
-
FERM and PDZ domain-containing protein 4
- S-SCAM:
-
membrane-associated guanylate kinase, WW and PDZ domaincontaining protein 2
- SHANK:
-
SH3 and multiple ankyrin repeat domains protein 1 (2, 3)
- SP:
-
spectrin α chain, non-erythrocytic 1
- DLG1:
-
Disks large homolog 1
- KL:
-
Kalirin
- α-Pix:
-
Rho guanine nucleotide exchange factor 6
- β-Pix:
-
Rho guanine nucleotide exchange factor 7
- DBNL:
-
Drebrin-like protein
- BAIAP2:
-
brainspecific angiogenesis inhibitor 1-associated protein 2
- Eph:
-
endophilin A3
References
Kjelstrup K.B., Solstad T., Brun V.H., Hafting T., Leutgeb S., Witter M.P., Moser E.I., Moser M.B. 2008. Finite scale of spatial representation in the hippocampus. Science. 321, 140–143. doi 10.1126/science.1157086
Hawley D.F., Morch K., Christie B.R., Leasure J.L. 2012. Differential response of hippocampal subregions to stress and learning. PLoS ONE. 7, e53126.
Bliss T.V., Collingridge G.L. 1993. A synaptic model of memory: Long-term potentiation in the hippocampus. Nature. 361, 31–39.
Ananko E.A., Podkolodny N.L., Stepanenko I.L., Podkolodnaya O.A., Rasskazov D.A., Miginsky D.S., Likhoshvai V.A., Ratushny A.V., Podkolodnaya N.N., Kolchanov N.A. 2005. GeneNet in 2005. Nucleic Acids Res. 33, 425–427.
Kolchanov N.A., Voevoda M.I., Kuznetsova T.N., Mordvinov V.A., Ignat’eva E.V. 2006. Gene networks of lipid metabolism. Byull. Sib. Otd. Ross. Akad. Med. Nauk. 2, 29–42.
Proskura A.L., Malakhin I.A., Turnaev I.I., Suslov V.V., Zapara T.A., Ratushnyak A.S. 2013. Intermolecular interactions in functional systems of neuron. Vavilov. Zh. Genet. Selekts. 17, 620–628.
Proskura A.L., Ratushnyak A.S., Zapara T.A. 2014. The protein–protein interaction networks of dendritic spines in the early phase of long-term potentiation. J. Comput. Sci. Syst. Biol. 7, 40–44.
Park M., Penick E.C., Edwards J.G., Kauer J.A., Ehlers M.D. 2004. Recycling endosomes supply AMPA receptors for LTP. Science. 305, 1972–1975.
Newpher T.M., Ehlers M.D. 2008. Glutamate receptor dynamics in dendritic microdomains. Neuron. 58, 472–497.
Shepherd J.D., Huganir R.L. 2007. The cell biology of synaptic plasticity: AMPA receptor trafficking. Annu. Rev. Cell Dev. Biol. 23, 613–643.
Patterson M.A., Szatmari E.M., Yasuda R. 2010. AMPA receptors are exocytosed in stimulated spines and adjacent dendrites in a Ras-ERK-dependent manner during long-term potentiation. Proc. Natl. Acad. Sci. U. S. A. 107, 15951–15956.
Tanaka H., Hirano T. 2012. Visualization of subunitspecific delivery of glutamate receptors to postsynaptic membrane during hippocampal long-term potentiation. Cell. Rep. 1, 291–298.
van der Sluijs P., Hoogenraad C.C. 2011. New insights in endosomal dynamics and AMPA receptor trafficking. Semin. Cell. Dev. Biol. 22, 499–505.
Malakhin I.A., Proskura A.L., Zapara T.A., Ratushnyak A.S. 2012. Effect of transport vesicles assembly to preserve the effectiveness of synaptic transmission. Vestn. Novosibirsk. Gos. Univ. 10, 14–20.
Folstein S.E., Folstein M.F. 1983. Psychiatric features of Huntington’s disease: Recent approaches and findings. Psychiatr. Dev. 1, 193–205.
Cummings J.L., Cunningham K. 1992. Obsessivecompulsive disorder in Huntington’s disease. Biol. Psychiatry. 31, 263–270.
Rosenblatt A. 2007. Neuropsychiatry of Huntington’s disease. Dialogues Clin. Neurosci. 9, 191–197.
Julien C.L., Thompson J.C., Wild S., Yardumian P., Snowden J.S., Turner G., Craufurd D. 2007. Psychiatric disorders in preclinical Huntington’s disease. J. Neurol. Neurosurg. Psychiatry. 78, 939–943.
Cardoso F. 2009. Huntington disease and other choreas. Neurol. Clin. 27, 719–736.
Schilling G., Sharp A.H., Loev S.J., Wagster M.V., Li S.H., Stine O.C., Ross C.A. 1995. Expression of the Huntington’s disease (IT15) protein product in HDpatients. Hum. Mol. Genet. 4, 1365–1371.
Mangiarini L., Sathasivam K., Seller M., Cozens B., Harper A., Hetherington C., Lawton M., Trottier Y., Lehrach H., Davies S.W., Bates G.P. 1996. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 87, 493–506.
Sapp E., Schwarz C., Chase K., Bhide P.G., Young A.B., Penney J., Vonsattel J.P., Aronin N., DiFiglia M. 1997. Huntingtin localization in brains of normal and Huntington’s disease patients. Ann. Neurol. 42, 604–612.
Korzhova V.V., Artamonov D.N., Vlasova O.L., Becprozvannyi I.B. 2014. Huntington’s disease: Molecular and cellular bases of pathology. Zh. Vyssh. Nervn. Deyat. 64, 359–375.
Li X.J., Li S.H., Sharp A.H., Nucifora F.C., Schilling G., Lanahan A., Worley P., Snyder S.H., Ross C.A. 1995. A huntingtin-associated protein enriched in brain with implications for pathology. Nature. 378, 398–402.
Gómez-Tortosa E., MacDonald M.E., Friend J.C., Taylor S.A., Weiler L.J., Cupples L.A., Srinidhi J., Gusella J.F., Bird E.D., Vonsattel J.P., Myers R.H. 2001. Quantitative neuropathological changes in presymptomatic Huntington’s disease. Ann. Neurol. 49, 29–34.
Zuccato C., Valenza M., Cattaneo E. 2010. Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol. Rev. 90, 905–981. doi 10.1152/physrev.00041.2009
Murphy K.P., Carter R.J., Lione L.A., Mangiarini L., Mahal A., Bates G.P., Dunnett S.B., Morton A.J. 2000. Abnormal synaptic plasticity and impaired spatial cognition in mice transgenic for exon 1 of the human Huntington’s disease mutation. J. Neurosci. 20, 5115–5123.
Milnerwood A.J., Cummings D.M., Dallérac G.M., Brown J.Y., Vatsavayai S.C., Hirst M.C., Rezaie P., Murphy K.P. 2006. Early development of aberrant synaptic plasticity in a mouse model of Huntington’s disease. Hum. Mol. Genet. 15, 1690–1703.
Ciamei A., Morton A.J. 2009. Progressive imbalance in the interaction between spatial and procedural memory systems in the R6/2 mouse model of Huntington’s disease. Neurobiol. Learn. Mem. 92, 417–428. doi 10.1016/j.nlm.2009.06.002
Dallérac G.M., Cummings D.M., Hirst M.C., Milnerwood A.J., Murphy K.P. 2016. Changes in dopamine signalling do not underlie aberrant hippocampal plasticity in a mouse model of Huntington’s disease. Neuromol. Med. 18, 146–153.
Cattaneo E. 2003. Dysfunction of wild-type huntingtin in Huntington disease. News Physiol. Sci. 18, 34–37.
Van Raamsdonk J.M., Pearson J, Rogers D.A., Bissada N., Vogl A.W., Hayden M.R., Leavitt B.R. 2005. Loss of wild-type huntingtin influences motor dysfunction and survival in the YAC128 mouse model of Huntington disease. Hum. Mol. Genet. 14, 1379–1392.
Ehlers M.D. 2000. Reinsertion or degradation of AMPA receptors determined by activity-dependent endocytic sorting. Neuron. 28, 511–525.
Tardin C., Cognet L., Bats C., Lounis B., Choquet D. 2003. Direct imaging of lateral movements of AMPA receptors inside synapses. EMBO J. 22, 4656–4665.
Man H.Y., Ju W., Ahmadian G., Wang Y.T. 2000. Intracellular trafficking of AMPA receptors in synaptic plasticity. Cell Mol. Life Sci. 57, 1526–1534.
Petrini E.M., Lu J., Cognet L., Lounis B., Ehlers M.D., Choquet D. 2009. Endocytic trafficking and recycling maintain a pool of mobile surface AMPA receptors required for synaptic potentiation. Neuron. 63, 92–105.
Kalchman M.A., Koide H.B., McCutcheon K., Graham R.K., Nichol K., Nishiyama K., Kazemi-Esfarjani P., Lynn F.C., Wellington C., Metzler M., Goldberg Y.P., Kanazawa I., Gietz R.D., Hayden M.R. 1997. HIP1, a human homologue of S. cerevisiae Sla2p, interacts with membrane-associated huntingtin in the brain. Nat. Genet. 16, 44–53.
Mishra S.K., Agostinelli N.R., Brett T.J., Mizukami I., Ross T.S., Traub L.M. 2001. Clathrin- and AP-2-binding sites in HIP1 uncover a general assembly role for endocytic accessory proteins. J. Biol. Chem. 276, 46230–46236.
Waelter S., Scherzinger E., Hasenbank R., Nordhoff E., Lurz R., Goehler H., Gauss C., Sathasivam K., Bates G.P., Lehrach H., Wanker E.E. 2001. The huntingtin interacting protein HIP1 is a clathrin and alpha-adaptinbinding protein involved in receptor-mediated endocytosis. Hum Mol Genet. 10, 1807–1817.
Metzler M., Legendre-Guillemin V., Gan L., Chopra V., Kwok A., McPherson P.S., Hayden M.R. 2001. HIP1 functions in clathrin-mediated endocytosis through binding to clathrin and adaptor protein 2. J. Biol. Chem. 276, 39271–39276.
Yao P.J., Bushlin I., Petralia R.S. 2006. Partially overlapping distribution of epsin1 and HIP1 at the synapse: Analysis by immunoelectron microscopy. J. Comp. Neurol. 494, 368–379.
Metzler M., Li B., Gan L., Georgiou J., Gutekunst C.A., Wang Y., Torre E., Devon R.S., Oh R., Legendre-Guillemin V., Rich M., Alvarez C., Gertsenstein M., McPherson P.S., Nagy A., et al. 2003. Disruption of the endocytic protein HIP1 results in neurological deficits and decreased AMPA receptor trafficking. EMBO J. 22, 3254–3266.
Legendre-Guillemin V., Metzler M., Charbonneau M., Gan L., Chopra V., Philie J., Hayden M.R., McPherson P.S. 2002. HIP1 and HIP12 display differential binding to F-actin, AP2, and clathrin. Identification of a novel interaction with clathrin light chain. J. Biol. Chem. 277, 19897–19904.
Wilbur J.D., Chen C.Y., Manalo V., Hwang P.K., Fletterick R.J., Brodsky F.M. 2008. Actin binding by Hip1 (huntingtin-interacting protein 1. and Hip1R (Hip1- related protein) is regulated by clathrin light chain. J. Biol. Chem. 283, 32870–32879.
Wilbur J.D., Hwang P.K., Brodsky F.M., Fletterick R.J. 2010. Accommodation of structural rearrangements in the huntingtin-interacting protein 1 coiled-coil domain. Acta Crystallogr. D: Biol. Crystallogr. 66, 314–318.
Hussain N.K., Jenna S., Glogauer M., Quinn C.C., Wasiak S., Guipponi M., Antonarakis S.E., Kay B.K., Stossel T.P., Lamarche-Vane N., McPherson P.S. 2001. Endocytic protein intersectin-l regulates actin assembly via Cdc42 and N-WASP. Nat. Cell Biol. 3, 927–932.
Modregger J., DiProspero N.A., Charles V., Tagle D.A., Plomann M. 2002. PACSIN 1 interacts with huntingtin and is absent from synaptic varicosities in presymptomatic Huntington’s disease brains. Hum. Mol. Genet. 11, 2547–2558.
Kessels M.M., Qualmann B. 2004. The syndapin protein family: Linking membrane trafficking with the cytoskeleton. J. Cell Sci. 117, 3077–3086.
Goh S.L., Wang Q., Byrnes L.J., Sondermann H. 2012. Versatile membrane deformation potential of activated pacsin. PLoS ONE. 7, e51628. doi 10.1371/journal. pone.0051628
Quan A., Robinson P.J. 2013. Syndapin, a membrane remodelling and endocytic F-BAR protein. FEBS J. 280, 5198–5212.
Sittler A., Wä lter S., Wedemeyer N., Hasenbank R., Scherzinger E., Eickhoff H., Bates G.P., Lehrach H., Wanker E.E. 1998. SH3GL3 associates with the Huntingtin exon 1 protein and promotes the formation of polygln-containing protein aggregates. Mol. Cell. 2, 427–436.
Milosevic I., Giovedi S., Lou X., Raimondi A., Collesi C., Shen H., Paradise S., O’Toole E., Ferguson S., Cremona O., de Camilli P. 2011. Recruitment of endophilin to clathrin-coated pit necks is required for efficient vesicle uncoating after fission. Neuron. 72, 587–601.
Setou M., Seog D.H., Tanaka Y., Kanai Y., Takei Y., Kawagishi M., Hirokawa N. 2002. Glutamate-receptor-interacting protein GRIP1 directly steers kinesin to dendrites. Nature. 417, 83–87.
Li S.H., Li H., Torre E.R., Li X.J. 2000. Expression of huntingtin-associated protein-1 in neuronal cells implicates a role in neuritic growth. Mol. Cell Neurosci. 16, 168–183.
Rong J., McGuire J.R., Fang Z.H., Sheng G., Shin J.Y., Li S.H., Li. J. 2006. Regulation of intracellular trafficking of huntingtin-associated protein-1 is critical for TrkA protein levels and neurite outgrowth. J. Neurosci. 26, 6019–6030.
Ma B., Savas J.N., Yu M.S., Culver B.P., Chao M.V., Tanese N. 2011. Huntingtin mediates dendritic transport of β-actin mRNA in rat neurons. Sci Rep. 1, 140. doi 10.1038/srep00140
Mandal M., Wei J., Zhong P., Cheng J., Duffney L.J., Liu W., Yuen E.Y., Twelvetrees A.E., Li S., Li X.J., Kittler J.T., Yan Z. 2011. Impaired alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor trafficking and function by mutant huntingtin. J. Biol. Chem. 286, 33719–33728.
Qualmann B., Kelly R.B. 2000. Syndapin isoforms participate in receptor-mediated endocytosis and actin organization. J. Cell Biol. 148, 1047–1062.
Loebrich S. 2014. The role of F-actin in modulating clathrin-mediated endocytosis: Lessons from neurons in health and neuropsychiatric disorder. Commun. Integr. Biol. 7, e28740. doi 10.4161/cib.28740
Wegner A.M., Nebhan C.A., Hu L., Majumdar D., Meier K.M., Weaver A.M., Webb D.J. 2008. N-Wasp and the arp2/3 complex are critical regulators of actin in the development of dendritic spines and synapses. J. Biol. Chem. 283, 15912–15920.
Imarisio S., Carmichael J., Korolchuk V., Chen C.W., Saiki S., Rose C., Krishna G., Davies J.E., Ttofi E., Underwood B.R., Rubinsztein D.C. 2008. Huntington’s disease: From pathology and genetics to potential therapies. Biochem. J. 412, 191–209.
Li X.J., Li S.H., Sharp A.H., Nucifora F.C., Schilling G., Lanahan A., Worley P., Snyder S.H., Ross C.A. 1995. A huntingtin-associated protein enriched in brain with implications for pathology. Nature. 378, 398–402.
Li S.H., Hosseini S.H., Gutekunst C.A., Hersch S.M., Ferrante R.J., Li X.J. 1998. A human HAP1 homologue. Cloning, expression, and interaction with huntingtin. J. Biol. Chem. 273, 19220–19227.
Nasir J., Duan K., Nichol K., Engelender S., Ashworth R., Colomer V., Thomas S., Disteche C.M., Hayden M.R., Ross C.A. 1998. Gene structure and map location of the murine homolog of the Huntington-associated protein, Hap1. Mamm. Genome. 9, 565–570.
Nasir J., Lafuente M.J., Duan K., Colomer V., Engelender S., Ingersoll R., Margolis R.L., Ross C.A., Hayden M.R. 2000. Human huntingtin-associated protein (HAP-1) gene: Genomic organisation and an intragenic polymorphism. Gene. 254, 181–187.
Dragatsis I, Dietrich P, Zeitlin S. 2000. Expression of the Huntingtin-associated protein 1 gene in the developing and adult mouse. Neurosci. Lett. 282, 37–40.
Truant R., Atwal R., Burtnik A. 2006. Hypothesis: Huntingtin may function in membrane association and vesicular trafficking. Biochem Cell Biol. 84, 912–917.
Colin E., Zala D., Liot G., Rangone H., Borrell-Pagès M., Li X.J., Saudou F., Humbert S. 2008. Huntingtin phosphorylation acts as a molecular switch for anterograde/retrograde transport in neurons. EMBO J. 27, 2124–2134.
Hirokawa N., Noda Y., Tanaka Y., Niwa S. 2009. Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol. 10, 682–696.
Zala D., Colin E., Rangone H., Liot G., Humbert S., Saudou F. 2008. Phosphorylation of mutant huntingtin at S421 restores anterograde and retrograde transport in neurons. Hum. Mol. Genet. 17, 3837–3846. doi 10.1093/hmg/ddn281
Chowdhury S., Shepherd J.D., Okuno H., Lyford G., Petralia R.S., Plath N., Kuhl D., Huganir R.L., Worley P.F. 2006. Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron. 52, 445–459.
Gao Y.G., Yan X.Z., Song A.X., Chang Y.G., Gao X.C., Jiang N., Zhang Q., Hu H.Y. 2006. Structural insights into the specific binding of huntingtin proline-rich region with the SH3 and WW domains. Structure. 14, 1755–1765.
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © A.L. Proskura, S.O. Vechkapova, T.A. Zapara, A.S. Ratushniak, 2017, published in Molekulyarnaya Biologiya, 2017, Vol. 51, No. 4, pp. 734–742.
Rights and permissions
About this article
Cite this article
Proskura, A.L., Vechkapova, S.O., Zapara, T.A. et al. Protein–protein interactions of huntingtin in the hippocampus. Mol Biol 51, 647–653 (2017). https://doi.org/10.1134/S002689331704015X
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S002689331704015X