Abstract
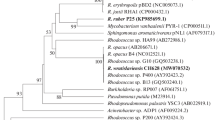
Polychlorinated biphenyls (PCBs) are persistent organic pollutants. Biphenyl 2,3-dioxygenase (BDO) is a key enzyme that determines the range of PCBs oxidized by a bacterial strain. BDO subunit α (BphA1) plays an essential role in substrate recognition and binding. The genes for dioxygenases that hydroxylate aromatic rings were screened and analyzed phylogenetically. Genes found in biphenyl-oxidizing Rhodococcus erythropolis strains G12a, B7b, and B106a proved to be similar to the published nucleotide sequences of the Rhodococcus sp. HA99 and R04 and Novosphingobium aromaticivorans F199 bphA1 genes, which code for the α-subunits that do not belong to the biphenyl/toluene dioxygenase (B/TDO) family. PCB-destructing R. ruber P25 was found to possess a unique bphA1 gene, which clusters together with the phenylpropionate dioxygenase (PPDO) α-subunits of Mycobacterium vanbaalenii PYR-1 and Frankia sp. EuI1c. The deduced amino acid sequences of the genes were analyzed. The amino acids of the BDO active site in R. wratislaviensis P1, P12, P13, and P20 (bphA1 genes of the B/TDO family) were identical to those of the active PCB degrader R. jostii RHA1. The Rhodococcus strains in question were shown to be active toward both orthoand parachlorinated ring of 2,4'-dichlorobiphenyl. The α-subunit amino acids responsible for the substrate specificity of the enzyme in Pseudomonas sp. S9, S13, S210, S211, and S212 (B/TDO family) were the same as in P. pseudoalcaligenes KF707. The Pseudomonas strains were active toward the para-chlorinated ring of 2,4'-dichlorobiphenyl. The results of screening bacterial strains for bphA1 can be used to identify the biotechnologically promising PCB destructors.
Similar content being viewed by others
Abbreviations
- BDO:
-
biphenyl dioxygenase
- NDO:
-
naphthalene dioxygenase
- PPDO:
-
phenylpropionate dioxygenase
- B/TDO:
-
biphenyl/toluene dioxygenase
- PCB:
-
polychlorinated biphenyl
References
Adams N.G., Richardson D.M. 1953. Isolation and identification of biphenyls from West Edmond Crude Oil. Anal. Chem. 25, 1073–1074.
http://www.unep/org
Abraham W.R., Nogales B., Golyshin P.N., Pieper D.H., Timmis K.N. 2002. Polychlorinated biphenyl-degrading microbial communities in soils and sediments. Curr. Opin. Microbiol. 5, 246–253.
Suenaga H., Watanabe T., Sato M., Ngadiman, Furukawa K. 2002. Alteration of regiospecificity in biphenyl dioxygenase by active-site engineering. J. Bacteriol. 184, 3682–3688.
Pieper D.H. 2005. Aerobic degradation of polychlorinated biphenyls. Appl. Microbiol. Biotechnol. 67, 170–191.
Parales R.E., Resnick S.M. 2004. Aromatic ring hydroxylating dioxygenases. In: Biodegradation and Bioremediation. Eds. Singh A., Ward O.P. Berlin: Springer, pp. 175–195.
Kumar P., Gómez- Gil L., Mohammadi M., Sylvestre M., Eltis L.D., Bolin J.T. 2011. Anaerobic crystallization and initial X-ray diffraction data of biphenyl 2,3-dioxygenase from Burkholderia xenovorans LB400: Addition of agarose improved the quality of the crystals. Acta Crystallogr. F: Struct. Biol. Cryst. Commun. 67, 59–62.
Kumar P., Mohammadi M., Dhindwal S., Pham T.T., Bolin J.T., Sylvestre M. 2012. Structural insights into the metabolism of 2-chlorodibenzofuran by an evolved biphenyl dioxygenase. Biochem. Biophys. Res. Commun. 421, 757–762.
Mohammadi M., Viger J.F., Kumar P., Barriault D., Bolin J.T., Sylvestre M. 2011. Retuning Rieske-type oxygenases to expand substrate range. J. Biol. Chem. 286, 27612–27621.
Colbert C.L., Agar N.Y., Kumar P., Chakko M.N., Sinha S.C., Powlowski J.B., Eltis L.D., Bolin J.T. 2013. Structural characterization of Pandoraea pnomenusa B-356 biphenyl dioxygenase reveals features of potent polychlorinated biphenyl-degrading enzymes. PLOS ONE. 8, 52550.
Ferraro D.J., Brown E.N., Yu C.L., Parales R.E., Gibson D.T., Ramaswamy S. 2007. Structural investigations of the ferredoxin and terminal oxygenase components of the biphenyl 2,3-dioxygenase from Sphingobium yanoikuyae B1. BMC Struct. Biol. 7, 10.
Nagarajan V., Sakurai N., Kubota M., Nonaka T., Nagumo H., Takeda H., Nishizaki T., Masai E., Fukuda M., Mitsui Y., Senda T. 2003. Crystallization of the terminal oxygenase component of biphenyl dioxygenase derived from Rhodococcus sp. strain RHA1. Protein Pept. Lett. 10, 412–417.
Furusawa Y., Nagarajan V., Tanokura M., Masai E., Fukuda M., Senda T. 2004. Crystal structure of the terminal oxygenase component of biphenyl dioxygenase derived from Rhodococcus sp. strain RHA1. J. Mol. Biol. 342, 1041–1052.
Suenaga H., Goto M., Furukawa K. 2006. Active-site engineering of biphenyl dioxygenase: Effect of substituted amino acids on substrate specificity and regiospecificity. Appl. Microbiol. Biotechnol. 71, 168–176.
Gibson D.T., Parales R.E. 2000. Aromatic hydrocarbon dioxygenases in environmental biotechnology. Curr. Opin. Biotechnol. 11, 236–243.
Takeda H., Yamada A., Miyauchi K., Masai E., Fukuda M. 2004. Characterization of transcriptional regulatory genes for biphenyl degradation in Rhodococcus sp. strain RHA1. J. Bacteriol. 186, 2134–2146.
Iwasaki T., Miyauchi K., Masai E., Fukuda M. 2006. Multiple-subunit genes of the aromatic-ring-hudroxylating dioxygenase play an active role in biphenyl and polychlorinated biphenyl degradation in Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 72, 5396–5402.
Yang X., Liu X., Song L., Xie F., Zhang G., Qian S. 2007. Characterization and functional analysis of a novel gene cluster involved in biphenyl degradation in Rhodococcus sp. strain R04. J. Appl. Microbiol. 103, 2214–2224.
Taguchi K., Motoyama M., Iida T., Kudo T. 2007. Polychlorinated biphenyl/biphenyl degrading gene clusters in Rhodococcus sp. K37, HA99, and TA431 are different from well-known bph gene clusters of rhodococci. Biosci. Biotechnol. Biochem. 71, 1136–1144.
Demanèche S., Meyer C., Micoud J., Louwagie M., Willison J.C., Jouanneau Y. 2004. Identification and functional analysis of two aromatic-ring-hydroxylating dioxygenases from a sphingomonas strain that degrades various polycyclic aromatic hydrocarbons. Appl. Environ. Microbiol. 70, 6714–6725.
Iwai S., Chai B., Sul W.J., Cole J.R., Hashsham S.A., Tiedje J.M. 2010. Gene-targeted-metagenomics reveals extensive diversity of aromatic dioxygenase genes in the environment. ISME J. 4, 279–285.
Lee T.K., Lee J., Sul W.J., Iwai S., Chai B., Tiedje J.M., Park J. 2011. Novel biphenyl-oxidizing bacteria and dioxygenase genes from a Korean tidal mudflat. Appl. Environ. Microbiol. 77, 3888–3891.
Jadeja N.B., More R.P., Purohit H.J., Kapley A. 2014. Metagenomic analysis of oxygenases from activated sludge. Bioresour. Technol. 165, 250–256.
Lozada M., Riva Mercadal J.P., Guerrero L.D., Di Marzio W.D., Ferrero M.A., Dionisi H.M. 2008. Novel aromatic ring-hydroxylating dioxygenase genes from coastal marine sediments of Patagonia. BMC Microbiol. 8, 50.
Taylor P.M., Medd J.M., Schoenborn L., Hodgson B., Janssen P.H. 2002. Detection of known and novel genes encoding aromatic ring-hydroxylating dioxygenases in soils and in aromatic hydrocarbon-degrading bacteria. FEMS Microbiol. Lett. 216, 61–66.
Standfuss-Gabisch C., Al-Halbouni D., Hofer B. 2012. Characterization of biphenyl dioxygenase sequences and activities encoded by the metagenomes of highly polychlorobiphenyl-contaminated soils. Appl. Environ. Microbiol. 78, 2706–2715.
Witzig R., Junca H., Hecht H.-J., Pieper D.H. 2006. Assessment of toluene/biphenyl dioxygenase gene diversity in benzene-polluted soils: Links between benzene biodegradation and genes similar to those encoding isopropylbenzene dioxygenases. Appl. Environ. Microbiol. 72, 3504–3514.
Shumkova E.S., Egorova D.O., Korsakova E.S., Plotnikova E.G., Dorofeeva L.V. 2014. Molecular biological characterization of biphenyl-degrading bacteria and identification of the biphenyl 2,3-dioxygenase a-subunit genes. Microbiology (Moscow). 83, 160–168.
Plotnikova E.G., Rybkina D.O., Anan’ina L.N., Yastrebova O.V., Demakov V.A. 2006. Characteristics of microorganisms isolated from technogenic soils of the Kama region. Russ. J. Ecol. 37, 233–240.
Plotnikova E.G., Egorova D.O., Shumkova E.S., Solyanikova I.P., Golovleva L.A. 2012. Degradation of 4-chlorobiphenyl and 4-chlorobenzoic acid by the strain Rhodococcus ruber P25. Microbiology (Moscow). 81, 143–153.
Zaitsev M.G., Tsoi T.V., Grishenkov V.G., Plotnikova E.G., Boronin A.M. 1991. Genetic control of degradation of chlorinated benzoic acids in Arthrobacter globiformis, Corynebacterium sepedonicum, and Pseudomonas cepacia strains. FEMS Microbiol. Lett. 81, 171–176.
Maniatis, T., Fritsch, E.F., Sambrook, J. 1982. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press.
Ausbel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K. 1995. Short Protocols in Molecular Biology, 3rd ed. New York: Wiley.
Iwai S., Johnson T.A., Chai B., Hashsham S.A., Tiedje J.M. 2011. Comparison of the specificities and efficacies of primers for aromatic dioxygenase gene analysis of environmental samples. Appl. Environ. Microbiol. 77, 3551–3557.
Shumkova E.S., Plotnikova E.G. 2013. Testing of primers designed for biphenyl 2,3-dioxygenase a-subunit genes detection in bacteria isolated from contaminated soil. Bull. Perm Univ. 3, 59–64.
Anan’ina L.N., Yastrebova O.V., Demakov V.A., Plotnikova E.G. 2011. Naphthalene-degrading bacteria of the genus Rhodococcus from the Verkhnekamsk salt mining region of Russia. Antonie Van Leeuwenhoek. 100, 309–316.
Larkin M.J., Allen C.C., Kulakov L.A., Lipscomb D.A. 1999. Purification and characterization of a novel naphthalene dioxygenase from Rhodococcus sp. strain NCIMB12038. J. Bacteriol. 181, 6200–6204.
Barriault D., Sylvestre M. 1999. Catalytic activity of Pseudomonas putida strain G7 naphthalene 1,2-dioxygenase on biphenyl. Int. Biodeterior. Biodegrad. 44, 33–37.
Diaz E., Ferrandez A., Garcia J. 1998. Characterization of the hca cluster encoding the dioxygenolytic pathway for initial catabolism of 3-phenylpropionic acid in Escherichia coli K-12. J. Bacteriol. 180, 2915–2923.
Ohmori T., Morita H., Tanaka M., Miyauchi K., Kasai D., Furukawa K., Miyashita K., Ogawa N., Masai E., Fukuda M. 2011. Development of a strain for efficient degradation of polychlorinated biphenyls by patchwork assembly of degradation pathways. J. Biosci. Bioeng. 111, 437–442.
Furusawa Y., Nagarajan V., Tanokura M., Masai E., Fukuda M., Senda T. 2004. Crystal structure of the terminal oxygenase component of biphenyl dioxygenase derived from Rhodococcus sp. strain RHA1. J. Mol. Biol. 342, 1041–1052.
Zielinski M., Kahl S., Hecht H.J., Hofer B. 2003. Pinpointing biphenyl dioxygenase residues that are crucial for substrate interaction. J. Bacteriol. 185, 6976–6980.
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © E.S. Shumkova, D.O. Egorova, S.V. Boronnikova, E.G. Plotnikova, 2015, published in Molekulyarnaya Biologiya, 2015, Vol. 49, No. 4, pp. 638–648.
Rights and permissions
About this article
Cite this article
Shumkova, E.S., Egorova, D.O., Boronnikova, S.V. et al. Polymorphism of the bphA genes in bacteria destructing biphenyl/chlorinated biphenils. Mol Biol 49, 569–580 (2015). https://doi.org/10.1134/S0026893315040159
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893315040159