Abstract
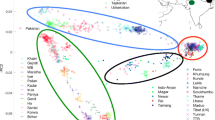
The gene pool structure of aboriginal Siberian populations has been described based on the polymorphism of the ZFX gene located on the chromosome X. In the ten populations studied, 49 haplotypes were present, three of them with high frequencies. Comparison of the obtained results with the available data from the HapMap project revealed unique African haplotypes that occurred in the Yoruba with the frequency of 3–7% and were not found in other populations. The genetic differentiation coefficient of the Siberian ethnic groups studied was 0.0486. Correlation analysis using Mantel’s test did not detect significant correlations between the genetic distance matrix and the matrices of geographic, linguistic, and anthropological differences, although the correlation with the anthropological matrix was the highest. Phylogenetic analysis proved strong isolation of the African population from the other ethnic groups investigated. The Siberian populations were divided into two separate clusters: the first one included Yakuts, Buryats, and Kets, while the second cluster included Altaians, Tuvinians, and Khanty. Using the principal component analysis, the populations were combined into three groups clearly differing by manifestation of Caucasoid and Mongoloid components. The first group included residents of Europe and one of Khanty populations, the second group included populations of South Siberia and residents of China. Mongoloid populations of East Siberia, the Japanese, and Kets were combined into the third group. Barrier analysis revealed a similar structure of genetic differentiation of Siberian populations. Linkage disequilibrium structure was obtained for six ethnic groups of Siberia. In five of them (except for the Ket population), ten ZFX SNPs formed a single linkage block.
Similar content being viewed by others
References
Xiao F.-X., Yotova V., Zietkiewicz E., et al. 2004. Human X-chromosomal lineages in Europe reveal Middle Eastern and Asiatic contacts. Eur. J. Hum. Genetics. 12, 301–311.
Laan M. 2005. X-chromosome as a marker for population history: Linkage disequilibrium and haplotype study in Eurasian populations. Eur. J. Hum. Genetics. 13, 452–462.
Tomas C., Juan J., Sanchez J.J., Barbaro A., Brandt-Casadevall C., Hernandez A., Ben Dhiab M., Ramon M., Morling N. 2008. X-chromosome SNP analyses in 11 human Mediterranean populations show a high overall genetic homogeneity except in North-west Africans (Moroccans). BMC Evol. Biol. 8, 1–14.
Casto A.M., Li J.Z., Absher D., Myers R., Ramachandran S., Feldman M.W. 2010. Characterization of X-Linked SNP genotypic variation in globally-distributed human populations. Genome Biol. 11, R10
Cann R. L., Stoneking M., Wilson A. C. 1987. Mitochondrial DNA and human evolution. Nature. 325, 31–36.
Lucotte G., Guerin P., Halle L., Loirat F., Hazout S. 1989. Y chromosome DNA polymorphisms in two African populations. Am. J. Hum. Genet. 45, 16–20.
Hey J. 1997. Mitochondrial and nuclear genes present conflicting portraits of human origins. Mol. Biol. Evol. 14, 166–172.
Zietkiewicz E. et al. 1997. Nuclear DNA diversity in worldwide distributed human populations. Gene. 205, 161–171.
Nachman M.W., Bauer V. L., Crowell S. L., Aquadro C.F. 1998. DNA variability and recombination rates at X-linked loci in humans. Genetics. 150, 1133–1141.
Jaruzelska J. et al. 1999. Spatial and temporal distribution of the neutral polymorphisms in the last ZFX intron: Analysis of the haplotype structure and genealogy. Genetics. 152, 1091–1101.
Harris E.E., Hey J. 1999. X chromosome evidence for ancient human histories. Proc. Natl. Acad. Sci. USA. 96, 3320–3324.
Safner S.F. 2004. The X chromosome in population genetics. Nature Rev. Genet. 5, 43–51.
Tishkoff S.A., Dietzsch E., Speed W., et al. 1996. Global patterns of linkage disequilibrium at the CD4 locus and modern human origins. Science. 271, 1380–1387.
Tishkoff S.A., Pakstis A.J., Stoneking M., et al. 2000. Short tandem-repeat polymorphism/Alu haplotype variation at the PLAT locus: Implications for modern human origins. Am. J. Hum. Genet. 67, 901–925
Kidd J.R., Pakstis A.J., Zhao H., et al. 2000. Haplotypes and linkage disequilibrium at the phenylalanine hydroxylase locus, PAH, in a global representation of populations. Am. J. Hum. Genet. 66, 1882–1899.
Reich D.E., Cargill M., Bolk S., Ireland J., Sabeti P.C., 2001. Linkage disequilibrium in the human genome. Nature. 411, 199–204.
Jorde L.B., Bamshad M., Rogers A.R. 1998. Using mitochondrial and nuclear DNA markers to reconstruct human evolution. Bioessays. 20, 126–136.
Stepanov V.A., Khitrinskaya I.Yu., Puzyrev V.P. 2001. Genetic differentiation of the Tuva population with respect to the Alu insertions. Genetika. 37, 563–569.
Khitrinskaya I.Yu., Stepanov V.A., Puzyrev V.P. 2001. Analysis of the Alu polymorphism in Buryat populations. Genetika. 37, 1553–1558.
Khitrinskaya I. Yu., Stepanov V. A., Puzyrev V. P., Spiridonova M. G., Puzyrev K. V.,. Maksimova N. R., and Nogovitsyna A. N. 2003. Genetic peculiarity of the Yakut population as inferred from autosomal loci. Mol. Biol. 37, 205–209.
Stepanov V.A. 2004. Genetic diversity and ethnogenesis of Tuvinians. Vestn. Etnich. Meditsiny. 1, 22–27.
Kharkov V.N., Stepanov V.A., Medvedeva O.F., Spiridonova M.G., Voevoda M.I., Tadinova V.N., Puzyrev V.P. 2007. Gene pool differences between northern and southern Altaians inferred from the data on Y-chromosomal haplogroups. Genetika. 43, 675–687.
Kharkov V.N., Stepanov V.A., Medvedeva O.F., Spiridonova M.G., Maksimova N.R., Nogovitsina A.N., Puzyrev V. P. 2008. The origin of Yakuts: Analysis of the Y-chromosome haplotypes. Mol. Biol. 42, 198–208.
Mantel N. 1967. The detection of disease clustering and a generalized regression approach. Cancer Res. 27, 209–220.
Nei M. 1987. Molecular Evolutionary Genetics. New York: Columbia Univ. Press.
Monmonier M. 1973. Maximum-difference barriers: An alternative numerical regionalization method. Geogr. Anal. 3, 245–261.
Manni F., Guerard E., Heyer E. 2004. Geographic patterns of (genetic, morphologic, linguistic) variation: How barriers can be detected by using Monmonier’s algorithm. Hum. Biol. 76, 173–190.
Barrett J.C., Fry B., Maller J., Daly M.J. 2005. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 21, 263–265.
Stepanov V.A. 2002. Ethnogenomics of Siberian and Central Asian populations. Med. Genet. 1, 113–123.
Schneider S., Roessli D., Excoffier L. 2000. Arlequin: A Software for population Genetics Data Analysis. Computer Program ver. 2.000. Geneva: Genetics and Biometry Lab, Dept. of Anthropology, Univ. of Geneva.
Brooks S.I. 1986. The World Population Today: Ethnodemographic Processes. Longon: Longman.
Roginskii Ya.Ya., Levin M.G. 1978. Antropologiya (Anthropology). Moscow: Nauka.
Narody mira. Istoriko-etnograficheskii spravochnik (Peoples of the World: Historical-Ethnographic Handbook). 1988 Moscow: Sovetskaya Entsiklopediya.
Narody zapadnoi Sibiri: Khanty. Mansi. Sel’kupy. Nentsy. Entsy. Nganasany. Kety (Peoples of Western Sibetua: The Khanty, Mansi, Selkups, Nentsy, Entsy, Hganasans, and Kets). 2005. Eds. Gemuev I.N., Molodin V.I., Sokolova Z.P. Moscow: Nauka.
Buryaty (The Buryats). 2004. Eds. Abaeva L.L., Zhukovskaya N.L. Moscow: Nauka.
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © I.Yu. Khitrinskaya, V.N. Khar’kov, V.A. Stepanov, 2010, published in Molekulyarnaya Biologiya, 2010, Vol. 44, No. 5, pp. 804–815.
Rights and permissions
About this article
Cite this article
Khitrinskaya, I.Y., Khar’kov, V.N. & Stepanov, V.A. Genetic diversity of the chromosome X in aboriginal Siberian populations: The structure of linkage disequilibrium and haplotype phylogeography of the ZFX locus. Mol Biol 44, 709–719 (2010). https://doi.org/10.1134/S0026893310050055
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893310050055