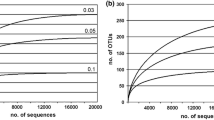
Abstract
Investigations into the microbial community of bottom sediments in the karst Lake Black Kichier were conducted. These sediments exhibited elevated levels of sulfide, dissolved methane, and organic matter. Direct radiotracer experiments revealed substantial rates of microbial processes involved in the decomposition of organic matter. Uncultivated archaea belonging to the phylum Thermoplasmatota were identified within the microbial community. Metagenomic analysis unveiled representatives from five orders: Methanomassiliicoccales, Thermoprofundales (formerly known as Marine Benthic Group D and DHVEG-1), DTX01, SG8-5, and Candidatus Gimiplasmatales (formerly UBA10834). These archaea were previously believed to occur exclusively in deep marine sediments characterized by extreme organic matter scarcity. This discovery reshapes our understanding of the role played by Thermoplasmatota archaea, spanning five orders, in the degradation segment of the carbon cycle.


Similar content being viewed by others
REFERENCES
Alneberg, J., Bjarnason, B.S., De Bruijn, I., Schirmer, M., Quick, J., Ijaz, U.Z., Lahti, L., Loman, N.J., Andersson, A.F., and Quince, C., Binning metagenomic contigs by coverage and composition, Nat. Methods, 2014, vol. 11, pp. 1144–1146.
Baker, B.J., De Anda, V., Seitz, K.W., Dombrowski, N., Santoro, A.E., and Lloyd, K.G., Diversity, ecology, and evolution of Archaea, Nat. Microbiol., 2020, vol. 5, pp. 887–900. https://doi.org/10.1038/s41564-020-0715-z
Borrel, G., Adam, P.S., and Gribaldo, S., Methanogenesis and the Wood–Ljungdahl pathway: an ancient, versatile, and fragile association, Genome Biol. Evol., 2016, vol. 8, pp. 1706–1711.
Capone, D.G. and Kiene, R.P., Comparison of microbial dynamics in marine and freshwater sediments—contrasts in anaerobic carbon catabolism, Limnol. Oceanogr., 1988, vol. 33, pp. 725–749.
Dong, X., Greening, C., Rattray, J.E., Chakraborty, A., Chuvochina, M., Mayumi, D., Dolfing, J., Li, C., Brooks, J.M., Bernard, B.B., Groves, R.A., Lewis, I.A., and Hubert, C.R.J., Metabolic potential of uncultured bacteria and archaea associated with petroleum seepage in deep-sea sediments. Nature Comm., 2019, vol. 10, no. 1, p. 1816. https://doi.org/10.1038/s41467-019-09747-0
Gorbunov, M.Yu. and Umanskaya, M.V., Karst Lakes of Mari Chodra National Park: stratification and vertical distribution of phototrophic plankton, 4th Conference on Actual Problems of Specially Protected Natural Areas, IOP Conf. Series: Earth and Environmental Science, 2020, vol. 607, p. 012019. https://doi.org/10.1088/1755-1315/607/1/012019
Gorlenko, V.M., Dubinina, G.A., and Kuznetsov, S.I., The Ecology of Aquatic Microorganisms, Stuttgart: E. Schweizerbartsche Verlagsbuchhandlung, 1983.
Han, X., Schubert, C.J., Fiskal, A., Dubois, N., and Lever, M.A., Eutrophication as a driver of microbial community structure in Lake sediments, Environ. Microbiol., 2020, vol. 22. pp. 3446–3462.
Hu, W., Pan, J., Wang, B., Guo, J., Li, M., and Xu, M., Metagenomic insights into the metabolism and evolution of a new Thermoplasmata order (Candidatus Gimiplasmatales), Environ. Microbiol., 2021, vol. 23, pp. 3695–3709.
Kadnikov, V.V., Mardanov, A.V., Beletsky, A.V., Banks, D., Pimenov, N.V., Frank, Y.A., Karnachuk, O.V., and Ravin, N.V., A metagenomic window into the 2-km-deep terrestrial subsurface aquifer revealed multiple pathways of organic matter decomposition, FEMS Microbiol. Ecol., 2018, vol. 94, p. fiy152.
Kadnikov, V.V., Savvichev, A.S., Mardanov, A.V., Beletsky, A.V., Merkel, A.Y., Ravin, N.V., and Pimenov, N.V., Microbial communities involved in the methane cycle in the near-bottom water layer and sediments of the meromictic subarctic Lake Svetloe, Antonie van Leeuwenhoek, 2019, vol. 112, pp. 1801–1814. https://doi.org/10.1007/s10482-019-01308-1
Kuznetsov, S.I., Mikroflora ozer i ee geokhimicheskaya deyatel’nost’ (Lake Microflora and Its Geochemichal Activity), Leningrad: Nauka, 1970.
Kuznezow, S.I. and Gorlenko, V.M., Limnologische und Mikrobiologische Eigenschaften von Karstseen der A.S.R. Mari, Arch. Hydrobiol., 1973, vol. 71, no. 4, pp. 475–486.
Parks, D.H., Chuvochina, M., Waite, D.W., Rinke, C., Skarshewski, A., Chaumeil, P.A., and Hugenholtz, P., A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life, Nat. Biotechnol., 2018, vol. 36, pp. 996–1004.
Parks, D.H., Imelfort, M., Skennerton, C.T., Hugenholtz, P., and Tyson, G.W., CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes, Genome Res., 2015, vol. 25, pp. 1043–1055.
Pimenov, N.V. and Bonch-Osmolovskaya, E.A., In situ activity studies in thermal environments, in Extremophiles. Methods in Microbiology, Rainey, F.A. and Oren, A., Eds., Elsevier, Academic Press, 2006, vol. 35, pp. 29–53.
Rinke, C., Rubino, F., Messer, L.F., Youssef, N., Parks, D.H., Chuvochina, M., Brown, M., Jeffries, T., Tyson, G.W., Seymour, J.R., and Hugenholtz, P., A phylogenomic and ecological analysis of the globally abundant Marine Group II archaea (Ca. Poseidoniales ord. nov.), ISME J., 2019, vol. 13, pp. 663–675. https://doi.org/10.1038/s41396-018-0282-y
Savvichev, A.S., Rusanov, I.I., Rogozin, D.Yu., Zakharova, E.E., Lunina, O.N., Bryantseva, I.A., Yusupov, S.K., Pimenov, N.V., Degermendzhi, A.G., and Ivanov, M.V., Microbiological and isotopic geochemical investigations of meromictic Lakes in Khakasia in winter, Microbiology (Moscow), 2005, vol. 74, no. 4, pp. 477–486.
Savvichev, A.S., Lunina, O.N., Rusanov, I.I., Zakharova, E.E., Veslopolova, E.F., and Ivanov, M.V., Microbiological and isotopic geochemical investigation of Lake Kislo-Sladkoe, a meromictic water body at the Kandalaksha Bay shore (White Sea), Microbiology (Moscow), 2014, vol. 83, pp. 56–66.
Savvichev, A., Rusanov, I., Dvornikov, Y., Kadnikov, V., Kallistova, A., Veslopolova, E., Chetverova, A., Leibman, M., Sigalevich, P., Pimenov, N., Ravin, N., and Khomutov, A., The water column of the Yamal tundra Lakes as a microbial filter preventing methane emission, Biogeosciences, 2021, vol. 18, pp. 2791–2807. https://doi.org/10.5194/bg-18-2791
Zemskaya, T.I., Bukin, S., Lomakina, A.V., and Pavlova, O.N., Microorganisms in the sediments of Lake Baikal, the deepest and oldest Lake in the world, Microbiology (Moscow), 2021, vol. 90, pp. 298–313.
Zheng, P.F., Wei, Z., Zhou, Y., Li, Q., Qi, Z., Diao, X., and Wang, Y., Genomic evidence for the recycling of complex organic carbon by novel Thermoplasmatota clades in deep-sea sediments, mSystems, 2022, vol. 7, p. e00077-22.
Zhou, Z., Liu, Y., Lloyd, K.G., Pan, J., Yang, Y., Gu, J.-D., and Li, M., Genomic and transcriptomic insights into the ecology and metabolism of benthic archaeal cosmopolitan, Thermoprofundales (MBG-D archaea). ISME J., 2019, vol. 13, pp. 885–901. https://doi.org/10.1038/s41396-018-0321-8
ACKNOWLEDGMENTS
The authors are grateful to the staff of the Kichiyer Sanatorium (Republic of Mari El) for organizational assistance in setting up a temporary laboratory.
Funding
Field works, sample processing, and molecular-biological data analysis performed by researchers of the Federal Research Center of Biotechnology of the Russian Academy of Sciences were supported by the Russian Science Foundation, project no. 22-14-00038. Radiotracer experiments were performed by I.I. Rusanov in the framework of State Assignment of the Ministry of Science and Higher Education.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This work does not contain any studies involving human or animal subjects.
CONFLICT OF INTEREST
The authors of this work declare that they have no conflict of interest.
Additional information
Translated by E. Makeeva
Publisher’s Note.
Pleiades Publishing remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kadnikov, V.V., Savvichev, A.S., Rusanov, I.I. et al. Metagenomic Analysis of Bottom Sediments of the Karst Meromictic Lake Black Kichier Revealed Abundant Unculturable Thermoplasmatota. Microbiology 93, 128–133 (2024). https://doi.org/10.1134/S0026261723604359
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026261723604359