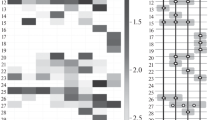
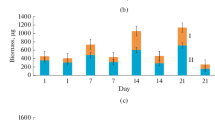
Abstract
Molecular biological techniques and bioinformatic analysis were used to investigate the phylogenetic and functional diversity of the prokaryotic complex of soil microcosms. The dominant organisms of the hydrolytic community were different in the samples from different climatic zones. In the soils subject to anthropogenic or abiogenic load, apart from decreased diversity and abundance of prokaryotes, the number of the genes marking the ability to degrade xenobiotics, as well as those encoding nitrogen conversion and metabolism of vitamins and cofactors, was found to increase. Under heavy oil contamination, the bacterial community was capable of nitrification; its role increased in the lower horizons of the soil profile. The patterns revealed in the work indicate high metabolic potential of the prokaryotic component of the studied soils.


Similar content being viewed by others
REFERENCES
Bürgmann, H., Widmer, F., Sigler, W.V., and Zeyer, J., mRNA extraction and reverse transcription-PCR protocol for detection of nifH gene expression by Azotobacter vinelandii in soil, Appl. Environ. Microbiol., 2003, vol. 69, pp. 1928–1935. https://doi.org/10.1128/AEM.69.4.1928-1935.2003
Hallin, S., Jones, C.M., Schloter, M., and Philippot, L., Relationship between n-cycling communities and ecosystem functioning in a 50-year-old fertilization experiment, ISME J., 2009, vol. 53, pp. 597–605. https://doi.org/10.1038/ismej.2008.128
Hendrickx, B., Junca, H., Vosahlova, J., Lindner, A., Ruegg, I., Bucheli-Witschel, M., Faber, F., Egli, T., Mau, M., Pieper, D.H., Top, E.M., Dejonghe, W., Bastiaens, L., and Springael, D., Alternative primer sets for PCR detection of genotypes involved in bacterial aerobic BTEX degradation: Distribution of the genes in BTEX degrading isolates and in subsurface soils of a BTEX contaminated industrial site, J. Microbiol. Methods, 2006, vol. 64, pp. 250–265. https://doi.org/10.1016/j.mimet.2005.04.018
Henry, S., Baudouin, E., López-Gutiérrez, J.C., Martin-Laurent, F., Brauman, A., and Philippot, L., Quantification of denitrifying bacteria in soils by nirK gene targeted real-time PCR, J. Microbiol. Methods, 2004, vol. 59, pp. 327–335. https://doi.org/10.1016/J.MIMET.2004.07.002
Gogmachadze, L.G., Khusnetdinova, K.A., Stepanov, A.L., and Kravchenko, I.K., Microcosm study of ammonium and drying impact on methane oxidation in agricultural soil, J. Agricult. Environ., 2023, vol. 36, no. 8, pp. 10–22. https://doi.org/10.23649/JAE.2023.36.7
Langille, M., Zaneveld, J., Caporaso, J.G., McDonald, D., Knights, D., Reyes, J., Clemente, J., Burkepile, D., Vega Thurber, R., Knight, R., Beiko, R., and Huttenhower, C., Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences, Nat. Biotechnol., 2013, vol. 31, no. 9, pp. 814–21. https://doi.org/10.1038/nbt.2676
Manucharova, N.A., Pozdnyakov, L.A., Vlasova, A.P., Yanovich, A.S., Ksenofontova, N.A., Kovalenko, M.A., Stepanov, P.Y., Gennadiev, A.N., Golovchenko, A.V., and Stepanov, A.L., Metabolically active prokaryotic complex in grassland and forests’ sod-podzol under polycyclic aromatic hydrocarbon influence, Forests (Basel), 202, vol. 12, no. 8, pp. 1103–1117. https://doi.org/10.3390/f12081103
Manucharova, N.A., Ksenofontova, N.A., Belov, A.A., Kamenskiy, N.N., Arzamazova, A.V., Zenova, G.M., Kinzhaev, R.R., Trofimov, S.Y., and Stepanov, A.L., Prokaryotic component of oil-contaminated oligotrophic peat soil under different levels of mineral nutrition: biomass, diversity, and activity, Euras. Soil Sci., 2021, vol. 54, no. 1, pp. 89–97. https://doi.org/10.31857/s0032180x2101010x
Markowitz, V.M., Chen, I.-M.A., Palaniappan, K., Chu, K., Szeto, E., Grechkin, Y., Ratner, A., Jacob, B., Huang, J., Williams, P., Huntemann, M., Anderson, I., Mavromatis, K., Ivanova, N.N., and Kyrpides, N.C., IMG: the Integrated Microbial Genomes database and comparative analysis system, Nucleic Acids Res., 2012, vol. 40. Database issue, pp. D115–122. https://doi.org/10.1093/nar/gkr1044
Samarghandi, M.R., Arabestani, M.R., Zafari, D., Rahmani, A.R., Afkhami, A., and Godini, K., Bioremediation of actual soil samples with high levels of crude oil using a bacterial consortium isolated from two polluted sites: investigation of the survival of the bacteria, Global NEST J., 2018, vol. 20, pp. 432–438.
Sutton, N.B., Maphosa, F., Morillo, J.A., Al-Soud, W.A., Langenhoff, A.A.M., Grotenhuis, T., Rijnaarts, H.H.M., and Smidt, H., Impact of long-term diesel contamination on soil microbial community structure, Appl. Environ. Microbiol., 2013, vol. 79, pp. 619–630.
Wang, Q., Garrity, G.M., Tiedje, J.M., and Cole, J.R., Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy, Appl. Environ. Microbiol., 2007, vol. 73, no. 16, pp. 5261–5267. https://doi.org/10.1128/AEM.00062-07
Wang, Q., Duan, B., Yang, R., Zhao, Y., and Zhang, L., Screening and identification of chitinolytic actinomycetes and study on the inhibitory activity against Turfgrass Rootrot disease fungi, J. Biosci. Medic., 2015, vol. 3, p. 56065. https://doi.org/10.4236/jbm.2015.33009
Funding
The composition of soil microbial communities was determined with financial support of the Russian Science Foundation, grant no. 21-14-00076.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This work does not contain any studies involving human and animal subjects.
CONFLICT OF INTEREST
The authors of this work declare that they have no conflicts of interest.
Additional information
Translated by D. Timchenko
Publisher’s Note.
Pleiades Publishing remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Manucharova, N.A., Vlasova, A.P., Kovalenko, M.A. et al. Biotechnological Potential of the Soil Microbiome. Microbiology 93, 145–148 (2024). https://doi.org/10.1134/S0026261723604335
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026261723604335