Abstract—
Bioaugmentation, i.e., increasing the abundance of certain microorganisms in the community by adding appropriate cells or establishing the conditions promoting their growth, is widely used in environmental technologies. Its application for launching of the anammox reactors is usually limited to introduction of anammox bacteria. We expected addition of nitrifiers during anammox bioreactor launching to stimulate the anammox process due to rapid production of nitrite, which anammox bacteria use for ammonium oxidation. The present work investigated the effect of introduction of a nitrifying community on the composition and activity of the microbial community in an anammox reactor. At the time of inoculation of a laboratory SBR reactor, an active nitrifying community (5 days old) (ASB) (bioaugmenting activated sludge, ASB) containing group I nitrifiers, primarily Nitrosospira, was added (1 : 100 by biomass) to anammox activated sludge (ASA) stored for 1 month at 4°C and exhibiting low metabolic activity. The use of ASB resulted in increased efficiency of nitrogen removal. While noticeable nitrogen removal in the control (7%) was observed since day 11 of incubation, nitrogen removal in the experimental reactor began on day 4 at the level of 20%. Nitrogen removal after 30 days of incubation was ~60% in the experiment and 20% in the control. The rate of ammonium oxidation in the presence of ASB increased due to activity of nitrifying bacteria (during the first 10 days of operation) and anammox bacteria of the genus Brоcadia, which were already present in ASA (throughout all period of operation). Activity of group II nitrifiers (genera Nitrobacter and Nitrococcus), which were present in ASB, prevented accumulation of nitrite, which in high concentrations is toxic to both nitrifiers and anammox bacteria. High activity of the Nitrosospira nitrifiers introduced with ASB probably provided the anammox bacteria with one of the substrates (nitrite), promoting their rapid growth. During subsequent operation of the reactor, nitrifiers of the genus Nitrosomonas from the initial ASA community were mainly responsible for growth of the anammox bacteria. Thus, ASA bioaugmentation at the loading of the anammox reactor by active nitrifiers resulted in significantly improved efficiency of ammonium removal via the anammox process and accelerated transition of the reactor to the working mode.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
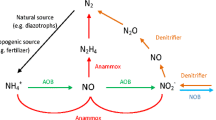
Biotechnological processes using the nitritation/anammox scheme (deammonification) have been actively used in recent years, since they are economically and ecologically more attractive than traditional processes based on the nitrification/denitrification scheme (Agraval et al., 2018; Kevbrina et al., 2019). Deammonification involves two stages: (1) approximately half of ammonium is oxidized to nitrite by ammonium-oxidizing bacteria (AOB) and (2) anammox bacteria (AnB) oxidize ammonium with nitrite to dinitrogen. The deammonification process requires relatively strict regulation of the physicochemical conditions to maintain the activity and competitiveness of AOB and AnB (Kallistova et al., 2016; Cho et al., 2020). This is especially the case for processing of concentrated return flows from facilities for processing wastewater sludge (methane fermentation) (Zhang et al., 2018; Izadi et al., 2021; Ochs et al., 2021; Pedrouso et al., 2021). The main task for research work in this area is to increase the activity and stress resistance of the major groups involved in deammonification, AOB and AnB, and to improve resistance of this process to unfavorable factors (Shourjeh et al., 2021).
One of the ways to achieve higher efficiency, quicker startup process, and stability of the biochemical parameters of wastewater treatment is bioaugmentation by adding to the reactor individual strains or mixed cultures in order to accelerate the transformation of pollutants (Herrero and Stuckey, 2015; Raper et al., 2018). Bioaugmentation with nitrifiers (Tang and Chen, 2015; Stenström and la Cour Jansen, 2017) or anammox bacteria (Jin et al., 2014; Zhang et al., 2018) was shown to enhance the efficiency of nitrification or the anammox process, respectively, as well as their stability under unfavorable conditions. Addition of AOB-enriched activated sludge to the bioreactor accelerated the onset of nitritation and established the conditions favoring the subsequent AnB development (Zhang et al., 2012; Ma et al., 2013). Nitrifiers are most often used for bioaugmentation of traditional nitrification/denitrification processes. The works on addition of nitrifiers to enhance the anammox process are few. Bioaugmentation with activated sludge enriched with AOB and AnB facilitated stabilization of the deammonification process and provided for more rapid commissioning of the bioreactors (Wett et al., 2013; Miao et al., 2017; Pedrouso et al., 2021). Ma et al. (2013) did not carry out AOB identification; activated sludge was added daily. Miao et al. (2017) achieved enrichment with the biomass of anammox bacteria by its monthly addition to the reactor.
The first stage (ammonium nitritation) is known to be the vulnerable point of the anammox process (Trojanowicz et al., 2021), since nitrifiers are sensitive to toxic compounds and rapidly die off in the course of storage or starvation (Salem et al., 2006). Anammox bacteria are considerably more robust, recover rapidly after periods of exposure to toxic agents, and die off slowly, not more than 1‒2% a day (Wang et al., 2018), compared to up to 20% a day for the nitrifiers (Salem et al., 2006).
Planning of the experiment was based on two prerequisites. Of two major bioaugmentation types, external (when the target biological agent is introduced into the system) and internal (when conditions are established for enrichment of the system with desired bacteria, i.e., selection), the former was chosen as the only one possible at launching of new reactors and during activity recovery in the reactor after poisoning of the activated sludge. Moreover, internal bioaugmentation is well-studied and is widely used, though often not termed bioaugmentation. This is the principle involved in the operation of skimmers enriching the activated sludge with phosphate-accumulating bacteria (Lema and Suarez, 2017) and partial nitrification reactors, enriching the sludge with ammonium-oxidizing bacteria (Tchobanoglous et al., 2014). Group I nitrifiers are required to support the activity of the anammox process, among which members of the genera Nitrosomonas and Nitrosospira predominate in the activated sludge. Addition of these bacteria was planned in the study.
The goal of the present work was to investigate the possibility of acceleration of the anammox process at launching the reactor by adding the physiologically active group I nitrifying bacteria.
MATERIALS AND METHODS
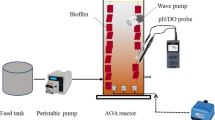
Cultivation of the activated sludge (anammox bacterial community) was carried out in the reactor described previously (Kallistova et al., 2020). The 4.5-L reactor consisted of two coaxial polymethyl methacrylate cylinders. The hermetic space between the cylinders was used for temperature control. Activated sludge was immobilized on a cylindrical carrier of fibrous plastic, a mixture of polyethylene and polypropylene (Polyvom, ETEK, Russia) with an internal diameter 85 mm, height 200 mm, and total surface area 11.6 dm2. Air was supplied from the bottom of the reactor by means of a SCHEGO SW2 compressor (Germany). When aeration was not applied, mixing was carried out using an IKA C-MAG MS7 magnetic stirrer (Germany) at 120‒150 rpm. The medium was supplied at the bottom of the reactor by Masterflex L/S economy drive peristaltic pumps (United States), and processed water was displaced through the upper fitting. The temperature mode was adjusted using an ELMI TV 2.03 water bath (Latvia) equipped with a circulation pump for the outer contour. The parameters were maintained with a Siemens LOGO 6ED1 programmed timer (Germany). The reactor was operated in the sequencing batch mode (SBR), which included the stages of settling, medium supply with the simultaneous removal of treated water, and aeration. The cycle duration was 6 h, and the average hydraulic time for the medium in the reactor was 27 h. The reactor operated at 30°C and oxygen concentrations of 0.4‒0.8 mg/L, with alternating phases of aeration and its absence (20 min each), and air flow of 20 L/h. Two identical reactors, the experimental and control ones, were used in the work. Oxygen concentration was determined with a WTW INOLAB 7310 meter equipped with a Sellox sensor (Germany).
The incoming medium contained the following (g/L): (NH4)2SO4, 0.942; NaCH3COO·3H2O, 0.04; KH2PO4, 0.044; NaHCO3, 2.1; pH 8.3 (Boeije et al., 1999). The medium was obtained by diluting the concentrate (prepared on distilled water) with tap water at the time of injection into the reactor. The anammox activated sludge (ASA) obtained from a previous run of the same reactor and stored for 30 days at 4°C was used as an inoculum. To launch the reactor, 1 L of the inoculum containing 2 g/L suspended matter was added to 3.5 L of the medium. The experimental reactor was supplemented with the association of nitrifying bacteria obtained as described below and containing 20 mg bacterial biomass; this corresponded to the 100 : 1 mass ratio of ASA and the nitrifying/bioaugmenting activated sludge (ASB). The concentrations of ammonium, nitrate, and nitrite ions were determined weekly in processed water using the standard techniques (Rice and Bridgewater, 2012). The amount of removed nitrogen (dN mg/L) was calculated as the difference between the concentration of ammonium nitrogen (N-NH4) in the inflowing medium and the total concentration of mineral nitrogen species in processed water (N-NH4, N-NO2, N-NO3). The efficiency of nitrogen removal was calculated as the share (%) of removed nitrogen to its concentration in supplied water.
Activity of ammonium-oxidizing bacteria (AOB), anammox bacteria (AnB), and nitrite-oxidizing bacteria (NOB) was calculated using the amount of removed nitrogen (dN) and the stoichiometry of the anammox process (Lotti et al., 2014):
Specific activity of AnB = dN/T, mg/L/h;
specific activity of AOB = ((N-\({\text{NH}}_{{4\,\,0}}^{ + }\) – N-\({\text{NH}}_{4}^{ + }\)) – (dN/1.99))/T, mg/L/h;
specific activity of NOB = (N-\({\text{NO}}_{{3\,\,{\text{r}}}}^{ - }\) – 0.08dN)/T, mg/L/h,
where T is hydraulic time of occurrence in the bioreactor, h;
N-\({\text{NH}}_{{4\,\,0}}^{ + }\) and N-\({\text{NH}}_{4}^{ + }\) are concentrations of ammonium nitrogen in the inflowing and processed medium, respectively, mg N/L,
N-\({\text{NO}}_{{3\,\,{\text{r}}}}^{ - }\) is the concentration of nitrate nitrogen in the effluent processed water.
The contribution of denitrification was not taken into account, since this process could be responsible for not more than 5% of nitrogen removal. Contribution to assimilation for the AnB growth was below 1% and was therefore also disregarded. The coefficients were taken from the work by Lotti et al. (2014).
Bioaumenting activated sludge (ASB) was a nitrifying enrichment culture obtained from composted mixture of excessive sludge from a processing plant of a dairy industry waste water and vegetable and wood waste in the 3 : 3 : 4 ratio (vol/vol). Compost samples were collected during the cooling stage at the temperature of 45°C from a 950-m3 industrial pile with the membranous opaque coverage and active aeration (Grunt Eko, Russia). Dry matter content in the compost (by mass) was 27.5 ± 1.3%. Water suspension of the compost was prepared by mixing 1 part of the compost with 9 parts (by mass) of the sterile nitrifier medium containing the following (g/L tap water): (NH4)2SO4, 2; K2HPO4·3H2O, 1; MgSO4·7H2O, 0.5; NaCl, 2; FeSO4·7H2O, 0.05; CaCO3, 5; pH 7. The suspension was mixed thoroughly for 10 min on a Vortex V-1 plus laboratory shaker (Biosan, Latvia). The enrichment culture was obtained by adding 5 mL of the compost suspension to 250 mL of the medium. The enrichment was cultured for 7 days at 30°C on a Biosan OS-20 incubator shaker (Biosan, Latvia) at 140 rpm. The culture was used as the inoculum (20%) to obtain the bioaugmenting culture (2 L), which was grown for 5 days under the same conditions and used as the bioaugmenting activated sludge (ASB) added to the anammox bioreactor. Cell titer in ASB (2 × 108 cells/mL) was determined using an Axio Imager M2 epifluorescence microscope (Carl Zeiss Microscopy, Germany).
The composition of the activated sludge microbial communities (in the inoculum, bioaugmenting ASB, and those immobilized on the carrier) was analyzed by high-throughput sequencing of the 16S rRNA gene fragments. Activated sludge was samples at the time of inoculation and after 25, 45, and 60 days of incubation.
Metagenomic DNA from the samples was isolated using the DNeasy PowerSoil Kit (Qiagen, Germany) according to the manufacturer’s protocols. The V3‒V4 variable region of the 16S rRNA gene was amplified using the universal primers 341F CCTAYGGGDBGCWSCAG and 806R GGACTACNVGGGTHTCTAAT (Frey et al., 2016). The amplicon libraries with barcodes were prepared from the obtained PCR fragments using the Nextera XT Index Kit v2 (Illumina, United States) according to the manufacturer’s protocols. PCR fragments were sequenced using Illumina MiSeq in the 2 × 300 nt format.
Paired reads were combined with FLASH v. 1.2.11 (Magoc and Salzberg, 2011). Low-quality reads, singletons, and chimeras were excluded at the next stage of analysis.
The remaining reads were clustered into operational taxonomic units (OTUs) with at least 97% sequence identity. To determine the share of an OTU in each sample, original reads (including the low-quality and singleton ones) were overlaid over the representative OTU sequences using the USEARCH v. 11 software package (Edgar, 2010). Taxonomic identification of the OTUs was carried out using the VSEARCH v. 2.14.1 algorithm and the Silva v. 138 database (Rognes et al., 2016).
The interaction between microbial groups was characterized using network analysis. Analysis of simultaneous OTU presence or of their mutual exclusion was performed based on the Spearman correlational matrix (Langfelder et al., 2012) using only the significant correlation values (Barberan et al., 2014). The threshold value accepted for the correlation coefficients was 0.6, and the one for the corrected p values was 0.001 (Luo and Bhattacharya, 2006). Only the OTUs with the relative abundance of at least 2.0% of the 16S rRNA gene sequences in at least one sample were included in the analysis. The networks were visualized using Cytoscape v. 3.8.2 (Shannon et al., 2003; Faust et al., 2016).
The 16S rRNA gene sequences obtained in the work were deposited to the NCBI database and are available at BioProject PRJNA556270.
Statistical processing of the data. In the experiments on cultivation of the anammox community and assessment of activity of various bacterial physiological groups, three measurements of each parameter (concentrations of oxygen and of nitrogen species) were carried out. The arithmetical mean and the mean absolute deviation were calculated. This value corresponded to the experimental dispersion and did not exceed 3%. The values are presented on the graphs as average ± mean deviation.
RESULTS AND DISCUSSION
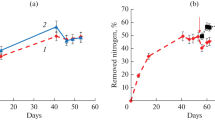
Parameters of bioreactor operation. Addition of ASB resulted in significantly more efficient removal of both ammonium and total mineral nitrogen from the bioreactor (Figs. 1, 2). On day 4, the concentrations of ammonium nitrogen in processed water decreased compared to the incoming water (with N-NH4 concentration 200 mg/L) by 40 and 14% in the bioreactors with and without (control) ASB bioaugmentation.
Noticeable removal of total nitrogen (7%) in the control reactor was observed from day 11 on; in the experimental reactor the process commenced on day 4 (20%). By the end of the experiment, ~60 and 20% of nitrogen was removed in the experimental and control reactors, respectively. During the first 10 days, nitrite concentrations in the experimental and control reactors did not differ considerably; subsequently, however, the concentration increased to 70‒75 mg/L in the control and decreased to 10‒15 mg/L in the experiment. Decreased concentrations of ammonium and nitrites, together with low concentrations of nitrate (and the absence of conditions for denitrification) indicated the key role of stage I nitrification and anammox reaction in the nitrogen balance.
The calculated values of specific activity for anammox bacteria, AOB, and NOB are shown on Fig. 3. Bioaugmentation resulted in increased nitrification activity at the beginning of the experiment. On day 4, AOB specific activity in the experimental variant was 1.8 times higher than in the control. At this time, the activity of anammox bacteria in the variant with bioaugmentation was 25 times higher than in the control. While AnB activity subsequently increased, even on day 32 specific activity of anammox bacteria in the experimental bioreactor was 2.7 times higher than in the control one. In both reactors activity of group II nitrifiers (NOB) was lower than the AOB activity by an order of magnitude. NOB activity in the presence of ASB was 5 times higher than in the control on day 4; during the next 2.5 weeks it rapidly decreased almost to zero. In the control activity of this group peaked on day 11 and then decreased to zero by day 18.
Composition of microbial communities. In order to determine the composition of the activated sludge microbial communities, 153 698 sequences of the V3‒V4 variable regions of the 16S rRNA genes were determined. Clusterization resulted in 1383 OTUs (Table S1). Alpha-diversity indices indicated high diversity of the communities in the original activated sludge samples (Table 1). In the course of operation, biodiversity in both the experimental and control bioreactors decreased and became similar (Table 1), which indicated selection of the microbial communities.
The OTUs found in the communities belonged to 22 bacterial phyla identified using GTDB (genome taxonomy database; Parks et al., 2018). Only seven of them were dominant and represented at least 1% of the 16S rRNA gene sequences in at least one sample (Fig. 4). Predominant groups belonged to the phyla Proteobacteria, Chloroflexi, Bacteroidota, Actinobacteriota, Planctomycetota, Verrucomicrobiota, and Spirochaetota; their total abundance in microbial communities exceeded 92%. Members of Acidobacteriota, Armatimonadota, Bdellovibrionota, Cyanobacteria, Deinococcota, Dependentiae, Desulfobacterota, Firmicutes, Gemmatimonadota, Hydrogenedentes, Myxococcota, Patescibacteria, Sumerlaeota, Synergistota, and WPS-2 were present as minor components, with their abundance not changing significantly in the course of community development. Archaea, belonging to the phylum Halobacterota, were found only in ASB, where their share was below 0.01%.
Relative abundance of bacterial phyla in the studied activated sludge samples: activated sludge of the control anammox bioreactor (ASA); activated sludge containing nitrifiers (ASB); and activated sludge from the experimental bioreactor supplemented with ASB (AB). The numerals after the sample designation indicate operational time in days.
ASB differed significantly from other samples, with predomination of Proteobacteria (the group to which nitrifiers belong), while the shares of Chloroflexi and Planctomycetota were low. Other activated sludge samples exhibited similar composition of microbial communities at the phylum level (Fig. 4).
Microorganism involved in nitrogen removal. The analyzed samples were found to contain nitrifiers of the genera Nitrosomonas (Otu8, Otu25, Otu359, Otu31, etc.) and Nitrosospira (mainly Otu2) belonging to the phylum Proteobacteria. In the course of cultivation, abundance of Nitrosomonas in the control AAS samples decreased from 7 to 4.5%, while in the ASB-supplemented bioreactor it increased from 5 to 10%. Interestingly, Nitrosospira were predominant nitrifiers in ASB (15.4%), while in the original AAS they were not present (Fig. 5). After addition of ASB (1 : 100 by biomass) to the experimental bioreactor, the share of Nitrosospira (Otu2) in the activated sludge decreased 100-fold (to 0.15%) and remained almost stable during subsequent cultivation (Table S1).
Relative abundance of nitrifiers and anammox bacteria in activated sludge samples: activated sludge from the anammox bioreactor (ASA); bioaugmented activated sludge with nitrifiers (ASB); and activated sludge from the anammox reactor supplemented with the bioaugmentinf sludge (AB). The numerals on the X axis indicate days of cultivation.
Detected anammox bacteria belonged to the genera Ca. Brocadia (Otu48 and Otu67) and Ca. Jettenia (Otu371). The share of Ca. Brocadia was significantly higher (7.8%); it decreased to 1.7% during operation of the bioreactor not supplemented with nitrifiers. In the bioreactor to which nitrifiers were added, the share of Ca. Brocadia remained almost stable (~6.5%). Relative abundance of Ca. Jettenia in all activated sludge samples decreased from 0.37 to 0.1%. Thus, nitrogen removal via the anammox reaction was probably carried out by Ca. Brocadia (Otu67); its relative abundance in the bioreactor supplemented with nitrifiers increased to 5.5%. Otu67 belonged to Ca. Brocadia fulgida (98.93% identity of the 16S rRNA gene sequences), which are common in anammox bioreactors. The second Ca. Brocadia phylotype, Otu48, belonged to another species, Ca. Brocadia caroliensis; its share decreased in the course of bioreactors operation.
OTU simultaneous presence and mutual exclusion. The functional characteristics of microorganisms are closely related to the properties of their habitats. Analysis of the presence of the major microbial groups in different microbial communities makes it possible to reveal the patterns of interaction between different microbial groups. We carried out network analysis of the studied microbial communities to reveal the groups most affected by the introduction of nitrifying bacteria. The analysis was carried out for the OTUs comprising over 2% in at least one sample. The results are presented on Fig. 6. It may be seen that Otu2 (Nitrospira) occurred together with Otu17 (Pedobacter) and Otu5 (uncultured Micavibrionales), which were dominant in ASB. However, no positive relations were found between Otu2 and the microorganisms of AAS.
It should be noted that the presence of anammox bacteria of Otu48 (Ca. Brocadia) correlated with the presence of nitrifiers Nitrosomonas (Otu8) and planctomycetes of the family Phycisphaeraceae (Otu50). Their relative abundance decreased in the course of operation of the anammox bioreactor.
Thus, ASB introduced into the microbial community had a significant effect on the efficiency of the anammox process, while the nitrifiers present in ASB could not adapt to the bioreactor conditions. Bacterial satellites introduced with ASB probably acted as the stimulators of the anammox process.
Increased efficiency of nitrogen removal correlated with increased abundance of members of the phyla Chloroflexi, Bacteroidetes, Leptospiraceae and Planctomycetes (Table S1). Negative correlation with abundance of Comamonas, Kapabacteria, Desulfuromonadia, Sorangiineae, Bdellovibrionia, Hyphomicrobium, and Chitinophagales was revealed. The reason for the negative correlation may be explained only for Bdellovibrionia, predatory bacteria capable of lysing other microbial cells. No functional relationships could be determined for other bacteria.
Another microbial group, addition of which could stimulate ammonium removal are the ASB bacteria, that preserved or increased their abundance after introduction into the bioreactor. In the initial AAS these organisms were not found or their share was minimal. These organisms belonged to Otu31 (Nitrosomonas), Otu125 (Lacunisphaera), Otu118 (uncultured planctomycete), Otu959 (Castellaniella_hirudinis), Otu66 (Castellaniella_ginsengisoli), Otu247 (Pedomicrobium), Otu383 (Sphingopyxis_sp.), Otu80 (Bordetella petrii), Otu1147 (Rhizobiales), Otu732 (Lacunisphaera), and Otu810 (Micavibrionales).
It should be noted that the share of stage II nitrifiers (nitrite oxidizers) was low in all activated sludge samples. Thus, ASB contained 0.01‒0.04% of this nitrifier type: Otu611 and Otu623 (Nitrobacter alkalicus) and Otu853 (Nitrococcus sp.). This finding was in agreement with their low activity (Fig. 3). Thus, both analysis of mineral nitrogen balance in the bioreactors and molecular biological investigation suggest the conclusion that the transformation of nitrogen compounds in the studied microbial communities followed mainly the “stage I nitrification‒anammox process” pathway. Both stage II nitrification and heterotrophic denitrification were insignificant.
ASB bioaugmentation with the Nitrosospira-enriched microbial community resulted in a rapid (in several hours‒days) increase of nitrification activity and anammox activity, with the latter subsequently maintained at the level several times higher than the anammox activity in the control bioreactor (without bioaugmentation). Increasing anammox activity correlated with increasing relative abundance of Ca. Brocadia fulgida (Otu67).
In general, our results confirmed the positive effect of bioaugmentation with nitrifiers, which has been used worldwide. However, bioaugmentation of wastewater treatment bioreactors is usually carried out with the material obtained on the spot and enriched with the microorganisms common in the activated sludge of a given waste treatment plant. Thus, Nitrosomonas predominates among stage I nitrifiers in activated sludge of industrial facilities (Agraval et al., 2018; Kevbrina et al., 2019), since it oxidizes ammonium more actively than Nitrosospira, the second most abundant group (Dytczak et al., 2007). Bioaugmentation with the material obtained on the spot is presently used at several large-scale plants. Bioaugmentation with nitrifying microorganisms from activated sludge is used in the BABE process, increasing the rate of nitrification in ammonium-enriched wastewater (Kallistova et al., 2016); a full-scale deammonification process was implemented in the main flow of the Strass Waste Water Treatment Plant (Strass, Austria) (Cho et al., 2020).
We performed bioaugmentation using the community enriched with Nitrosospira, bacteria less widespread in waste treatment plants, and obtained unexpected results. Although the nitrifying and anammox activities increased rapidly after bioaugmentation, in the course of subsequent operation Nitrosomonas (Otu25 and Otu359), belonging to the original community, grew more actively than the introduced Nitrosospira. This was probably due to the initially high activity of the introduced Nitrosospira nitrifiers, which provided the substrate (nitrite) for anammox bacteria and promoted a rapid increase in anammox activity. Nitrosospira was subsequently replaced by more competitive autochthonous nitrifiers.
REFERENCES
Agrawal, S., Seuntjens, D., Cocker, P.D., Lackner, S., and Vlaeminck, S., Success of mainstream partial nitritation/anammox demands integration of engineering, microbiome and modeling insights, Curr. Opin. Biotechnol., 2018, vol. 50, pp. 214–221.
Barberán, A., Ramirez, K.S., Leff, J.W., Bradford, M.A., Wall, D.H., and Fierer, N., Why are some microbes more ubiquitous than others? Predicting the habitat breadth of soil bacteria, Ecol. Lett., 2014, vol. 17, pp. 794–802. https://doi.org/10.1111/ele.12282
Cho, S., Kambey, C., and Nguyen, V.K., Performance of anammox processes for wastewater treatment: a critical review on effects of operational conditions and environmental stresses, Water, 2020, vol. 12, art. 20. https://doi.org/10.3390/w12010020
Dosta, J., Vila, J., Sancho, I., Basset, N., Grifoll, M., and Mata-Álvarez, J., Two-step partial nitritation/Anammox process in granulation reactors: start-up operation and microbial characterization, J. Environ. Manag., 2015, vol. 164, pp. 196–205.
Dytczak, M.A., Londry, K.L., and Oleszkiewicz, J.A., Activated sludge operational regime has significant impact on the type of nitrifying community and its nitrification rates, Water Res., 2008, vol. 42, pp. 2320–2328. https://doi.org/10.1016/j.watres.2007.12.018
Edgar, R.C., Search and clustering orders of magnitude faster than BLAST, Bioinform., 2010, vol. 26, pp. 2460–2461.
Faust, K. and Raes, J., CoNet app: inference of biological association networks using Cytoscape, F1000Res., 2016, vol. 5, p. 1519. https://doi.org/10.12688/f1000research.9050.2
Frey, B., Rime, T., Phillips, M., Stierli, B., Hajdas, I., Widmer, F., and Hartmann, M., Microbial diversity in European alpine permafrost and active layers, FEMS Microbiol. Ecol., 2016, vol. 92, art. fiw018. https://doi.org/10.1093/femsec/fiw018
Herrero, M. and Stuckey, D.C., Bioaugmentation and its application in wastewater treatment: a review, Chemosphere, 2015, vol. 140, pp. 119–128. https://doi.org/10.1016/j.chemosphere.2014.10.033
Izadi Parin, Izadi Parnian, and Eldyasti, A., Towards mainstream deammonification: comprehensive review on potential mainstream applications and developed sidestream technologies, J. Environ. Manag., 2021, vol. 279, art. 111615. https://doi.org/10.1016/j.jenvman.2020.111615
Jin, R.C., Zhang, Q.Q., Zhang, Z.Z., Liu, J.H., Yang, B.E., Guo, L.X., and Wang, H.Z., Bio-augmentation for mitigating the impact of transient oxytetracycline shock on anaerobic ammonium oxidation (ANAMMOX) performance, Bioresour. Technol., 2014, vol. 192, pp. 756–764.
Kallistova, A.Y., Nikolaev, Y.A., Berestovskaya, Y.Y., Grachev, V.A., Kostrikina, N.A., Pelevina, A.V., Pimenov, N.V., Mardanov, A.V., and Ravin, N.V., Investigation of formation and development of anammox biofilms by light, epifluorescence, and electron microscopy, Microbiology (Moscow), 2020, vol. 89, pp. 708‒719.
Kallistova, A.Y., Nikolaev, Y.A., Pimenov, N.V., Dorofeev, A.G., Kozlov, M.N., and Kevbrina, M.V., Role of anammox bacteria in removal of nitrogen compounds from wastewater, Microbiology (Moscow), 2016, vol. 85, pp. 140‒156.
Kevbrina, M.V., Dorofeev, A.G., Agerev, A.M., Kozlov, M.N., Nikolaev, Yu.A., and Aseeva, V.G., Anammox, a promising technology for nitrogen removal from wastewater, Vodosnab. San. Trhkh., 2019, no. 5, pp. 28–35.
Langfelder, P. and Horvath, S., Fast R functions for robust correlations and hierarchical clustering, J. Stat. Softw., 2012, vol. 46, art. i11.
Lema, J.M. and Suarez, S., Eds., Innovative Wastewater Treatment and Resource Recovery Technologies: Impacts on Energy, Economy and Environment, IWA, 2017. https://doi.org/10.2166/9781780407876
Lotti, T., Kleerebezem, R., Lubello, C., and van Loosdrecht, M.C.M., Physiological and kinetic characterization of a suspended cell anammox culture, Water Res., 2014, vol. 60, pp. 1–14.
Luo, X. and Bhattacharya, C.B., Corporate social responsibility, customer satisfaction, and market value, J. Mark., 2006, vol. 70, pp. 1–18.
Ma, B., Wang, S., Zhang, S., Li, X., Bao, P., and Peng, Y., Achieving nitritation and phosphorus removal in a continuous-flow anaerobic/oxic reactor through bio-augmentation, Bioresour. Technol., 2013, vol. 139, pp. 375‒378. https://doi.org/10.1016/j.biortech.2013.02.077
Magoc, T. and Salzberg, S., FLASH: Fast length adjustment of short reads to improve genome assemblies, Bioinform., 2011, vol. 27, pp. 2957–2963.
Miao, Y., Zhang, L., Li, B., Zhang, Q., Wang, S., and Peng, Y., Enhancing ammonium oxidizing bacteria activity was key to single-stage partial nitrification-anammox system treating low-strength sewage under intermittent aeration condition, Bioresour. Technol., 2017, vol. 231. P. 36–44. https://doi.org/10.1016/j.biortech.2017.01.045
Ochs, P., Martin, B.D., Germain, E., Stephenson, T., van Loosdrecht, M., and Soares, A., Ammonia removal from thermal hydrolysis dewatering liquors via three different deammonification technologies, Sci. Total Environ., 2021, vol. 755, part 1, art. 142684.
Parks, D.H., Chuvochina, M., Waite, D.W., Rinke, C., Skarshewski, A., Chaumeil, P.A., and Hugenholtz, P., A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life, Nat. Biotechnol., 2018, vol. 36, pp. 996–1004.
Pedrouso, A., Vázquez-Padín, J.R., Crutchik, D., and Campos, J.L., Application of Anammox-based processes in urban WWTPs: are we on the right track?, Processes, 2021, vol. 9, art. 1334. https://doi.org/10.3390/pr9081334
Raper E., Stephenson T., Anderson D.R., Fisherb R., Soares A. Industrial wastewater treatment through bioaugmentation, Proc. Saf. Environ. Prot. 2018, vol. 118, pp. 178–187. https://doi.org/10.1016/j.psep.2018.06.035
Rice, E.W. and Bridgewater, L., Eds., Standard Methods for the Examination of Water and Wastewater, Washington, DC: American Public Health Association, American Water Works Association, Water Environment Federation, 2012.
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F., VSEARCH: a versatile open source tool for metagenomics, PeerJ., 2016, October 18, e2584.
Salem, S., Berends, D.H.J.G., van der Roest, H.F., van der Kuji, R.J., and van Loosdrecht, M.C.M., Full-scale application of the BABE technology, Wat. Sci. Tech., 2003, vol. 50, pp. 87–96.
Salem, S., Moussa, M.S., and van Loosdrecht, M.C.M., Determination of the decay rate of nitrifying bacteria, Biotechnol. Bioeng., 2006, vol. 94, pp. 252–262. https://doi.org/10.1002/bit.20822
Shannon, P., Markiel, A., Ozier, O., Baliga, N.S., Wang, J.T., Ramage, D., Amin, N., Schwikowski, B., and Ideker, T., Cytoscape: a software environment for integrated models of biomolecular interaction networks, Genome Res., 2003, vol. 13. P. 2498‒2504. https://doi.org/10.1101/gr.1239303
Shourjeh, S.M., Kowal, P., Lu, X., Xie, L., and Drewnowski, J., Development of strategies for AOB and NOB competition supported by mathematical modeling in terms of successful deammonification implementation for energy-efficient WWTPs, Processes, 2021, vol. 9, art. 562. https://doi.org/10.3390/pr9030562
Stenström, F. and la Cour Jansen, J., Impact on nitrifiers of full-scale bioaugmentation, Water Sci. Technol., 2017, vol. 76, pp. 3079–3085. https://doi.org/10.2166/wst.2017.480
Tang, H.L. and Chen, H., Nitrification at full-scale municipal wastewater treatment plants: evaluation of inhibition and bioaugmentation of nitrifiers, Bioresour. Technol., 2015, vol. 190, pp. 76–81.
Tchobanoglous, G., Burton, F.L., and Stensel, H.D., Wastewater Engineering: Treatment and Reuse, Metcalf and Eddy, McGraw Hill, N.Y., 2014.
Trojanowicz, K., Trela, J., and Plaza, E., Possible mechanism of efficient mainstream partial nitritation/anammox (PN/A) in hybrid bioreactors (IFAS), Environ. Technol., 2021, vol. 42, pp. 1023‒1037. https://doi.org/10.1080/09593330.2019.1650834
Wang, Q., Song, K., Hao, X., Wei, J., Pijuan, M., van Loosdrecht, M.C.M., and Zhao, H., Evaluating death and activity decay of Anammox bacteria during anaerobic and aerobic starvation, Chemosphere, 2018, vol. 201, pp. 25–31. https://doi.org/10.1016/j.chemosphere.2018.02
Wett, B., Omari, A., Podmirseg, S., Han, M., Akintayo, O., Brandon, M.G., Murthy, S., Bott, C., Hell, M., and Takacs, I., Going for mainstream deammonification from bench to full scale for maximized resource efficiency, Water Sci. Technol., 2013, vol. 68, pp. 283–289.
Wett, B., Podmirseg, S.M., Gomez-Brandon, M., Hell, M., Nyhuis, G., Bott, C., and Murthy, S., Expanding DEMON sidestream deammonification technology towards mainstream application, Water Environ. Res., 2014, vol. 87, pp. 2084–2089.
Yang, Y., Zhang, L., Cheng, J., Zhang, S., Li, X., and Peng, Y., Microbial community evolution in partial nitritation/anammox process: from sidestream to mainstream, Bioresour. Technol., 2018, vol. 251, pp. 327–333.
Zhang, L., Zhang, S.J., Gan, Y.P., and Peng, Y.Z., Bio-augmentation to rapid realize partial nitrification of real sewage, Chemosphere, 2012, vol. 88, pp. 1097–1102.
Zhang, Q.-Q., Yang, G.-F., Sun, K.-K., Tian, G.-M., and Jin, R.-C., Insights into the effects of bio-augmentation on the granule-based anammox process under continuous oxytetracycline stress: performance and microflora structure, Chem. Engin. J., 2018, vol. 348, pp. 503–513. https://doi.org/10.1016/j.cej.2018.04.204
Zhang, Q., Vlaeminck, S.E., DeBarbadillo, C., Su, C., Al-Omari, A., Wett, B., Pümpel, T., Shaw, A., Chandran, K., Murthy, S., and De Clippeleira, H., Supernatant organics from anaerobic digestion after thermal hydrolysis cause direct and/or diffusional activity loss for nitritation and anammox, Water Res., 2018, vol. 143, pp. 270–281.
Funding
The work was supported by the Russian Foundation for Basic Research, project no. 18-29-08008 (operation of the anammox bioreactor), Russian Science Foundation, project no. 21-64-00019 (molecular analysis of microbial communities), and State Assignment for the Research Centre of Biotechnology, Russian Academy of Sciences (obtaining the nitrifying bacteria).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflicts of interest.
This article does not contain any studies involving animals or human participants performed by any of the authors.
Additional information
Translated by P. Sigalevich
Supplementary Information
Rights and permissions
Open Access. This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pimenov, N.V., Nikolaev, Y.A., Dorofeev, A.G. et al. Bioaugmentation of Anammox Activated Sludge with a Nitrifying Bacterial Community as a Way to Increase the Nitrogen Removal Efficiency. Microbiology 91, 133–142 (2022). https://doi.org/10.1134/S0026261722020102
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026261722020102