Abstract—
The taxonomic diversity and metabolic activity of microbial communities in the Laptev Sea water column above and outside the methane seep field were studied. The concentrations of dissolved methane in the water column at both stations were comparable until the depth of the pycnocline (25 m). At this depth, local methane maxima were recorded, with the highest concentration (116 nM CH4) found at the station outside the methane seep field. Results of the 16S rRNA gene sequencing and measurements of the rates of hydrogenotrophic methanogenesis indicated the absence of methanogenesis caused by the methanogenic archaea in the pycnocline and in other horizons of the water column. The 16S rRNA-based analysis of microbial phylogenetic diversity, as well as radiotracer analysis of the rates of primary production (PP), dark CO2 assimilation (DCA), and methane oxidation (MO), indicated the functioning of a diverse community of pelagic microorganisms capable of transforming a wide range of organic compounds under oligotrophic conditions of the Arctic basin. Hydrochemical prerequisites and possible microbial agents of aerobic methane production via demethylation of methylphosphonate and decomposition of dimethylsulfoniopropionate using dissolved organic matter synthesized in the PP, DCA, and MO processes are discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
The methane cycle is an important component of the carbon biogeochemical cycle in the ocean. Methane may either arrive to marine basins from deep hydrothermal springs, mud volcanoes, and cold methane seeps or be synthesized in situ in the sediments and the water column.
Methanogenic archaea, which are considered the main methane producers of our planet, are well-known and have been thoroughly studied. They are strict anaerobes and produce methane mainly from such low-molecular mass compounds as CO2, acetate, methanol, and methylamines. Methane formation from methoxy groups of cyclic organic compounds was recently shown (see review by Kurth et al., 2020 and references therein). Development of methanogenic archaea in the seas is associated mainly with anoxic sediments and (in the water column) with anoxic microniches in large suspended organic particles or in the zooplankton gut and pellets (Bianchi et al., 1992; Ditchfield et al., 2012). However, the surface, oxygen-containing waters of various areas of the Global Ocean often exhibit methane oversaturation relative to the atmosphere, the phenomenon termed the methane paradox (Karl et al., 2008; Bižic et al., 2020). Detection of anoxic microniches harboring methanogenic archaea has long been used to explain this phenomenon. The methane paradox is, however, presently explained as a result of metabolic activity of aerobic pelagic microorganisms, usually of methylphosphonate (MPn) degradation by aerobic bacteria under phosphate limitation (Karl et al., 2008; Sosa et al., 2019), which, apart from phosphate release, results in reduction of the methyl group to methane. Moreover, recent data indicate other mechanisms of aerobic methane production, although their specific features and occurrence in the Global Ocean remain poorly studied. Thus, under nitrogen limitation and available phosphorus, methane may be produced via aerobic decomposition of dimethylsulfoniopropionate (DMSP) to dimethyl sulfide (DMS) or methylmercaptan (methane thiol, MeSH) (Damm et al., 2010) with subsequent anaerobic degradation of these products by methylotrophic methanogens (Kurth et al., 2020). Moreover, recent studies revealed methane production by diverse phytoplanktonic microorganisms: cyanobacteria, coccolithophorids and other haptophytes, diatoms (see review by Bižić et al., 2020 and references therein). In this case, the issue is associated not with previously known metabolic pathways (MPn demethylation or production of hydrogen by nitrogenase and hydrogenases, with its subsequent utilization in hydrogenotrophic methanogenesis), but with an unknown mechanism of CO2 conversion to methane linked to the basic metabolism of oxygenic phototrophs.
The eastern Arctic shelf seas, including the Laptev Sea, are known as sources of massive methane emission to the atmosphere (Berchet et al., 2016; Shakhova et al., 2010). Over 700 sites of methane seepage with intensities varying by 5 orders of magnitude (up to 176 g CH4/(m2 day)), depending on the state of underwater permafrost and the stability of hydrates, have been revealed in the Laptev Sea (Shakhova et al., 2015). Concentrations of dissolved methane in near-bottom water at the zones of methane seep discharge are as high as 300–650 nМ and sometimes reach 5000 nМ (Shakhova et al., 2010; Savvichev et al., 2018). According to the long-term studies, methane transport from the sediments into the atmosphere occurs mainly via bubble transfer, which involves partial methane dissolution in the water column and its saturation (Shakhova et al., 2010, 2015). Elevated methane concentrations in the water columns of Arctic seas, including the Laptev Sea, are therefore usually associated with activity of numerous methane seeps. However, phylogenetic diversity and the metabolic potential of pelagic microbial communities of the Laptev Sea shelf that may act as alternative methane sources remain unstudied.
In order to assess the possible agents of microbial methane production under oxic conditions of the Laptev Sea water column, comparative study of the hydrochemical parameters was carried out in the present work, as well as determination of the taxonomic diversity and functional activity of pelagic microbial communities at stations located both above and outside the field of methane seeps.
MATERIALS AND METHODS
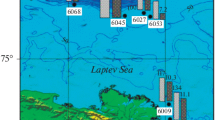
Research site and water sampling. Water and sediment samples were collected in the Laptev Sea during the 73rd cruise of RV Akademik Mstislav Keldysh in October 2018. Two stations of similar depth were chosen: AMK73-6045 (76.7744 N, 125.7620 E) in the zone of a methane seep discharge and AMK73-6053 (76.7387 N, 128.4512 E) outside the zone of methane bubble emission (Fig. 1). Water samples were collected from 10-L bathometers of a Rosette complex immediately after heaving on board. The sampling horizons were determined according to the data on temperature and salinity. The major parameters of the sampling stations are listed in Table 1.
Hydrochemical parameters of the water. Water samples were collected using transparent silicon rubber tubes in order to control the absence of gas bubbles. The samples for determination of dissolved oxygen and ammonium nitrogen were fixed immediately after collection. The samples for determination of phosphates and nitrates were collected into 0.5-L plastic vials without fixation and were analyzed in the laboratory on board.
Dissolved oxygen was determined using the modified Winkler method using calibrated 20–30-mL vials with 10 mm plug diameter and at least 300% washing volume (Methods.., 1999). Phosphates were determined by the Murphy-Riley method; nitrites were measured with the single color reagent; nitrates, according to the modified method of Morris and Riley; and ammonium nitrogen was measured according to the method of Sagi and Solórzano with formation of a phenol-hypochlorite complex (Methods.., 1999).
The N* quasiconservative indicator was calculated as: N* = (N – 16P + 2.90 µmol/kg) × 0.87 (Gruber and Sarmiento, 1997), where N is nitrate concentration and P is phosphate concentration. N* is close to zero when the Redfield proportion (N : P = 16 : 1) is maintained, while the positive and negative N* values indicate deviations from this ratio.
Samples for determination of methane concentrations were collected (avoiding turbulence) into glass vials with a fixing compound (KOH) and the standard headspace and sealed hermetically with rubber stoppers. Methane was determined by the phase equilibrium degassing method on a Krisrall-2000-M gas chromatograph (Chromatec, Yoshkar-Ola, Russia) equipped with a flame ionization detector. The error of this method was ±6%.
Radiotracer analysis. The rates of photosynthesis, microbial dark CO2 assimilation, and autotrophic methanogenesis were measured by the radiocarbon method with NaH14CO3; the rate of methane oxidation was determined using 14CH4.
To measure the rates of primary production (PP) and dark CO2 assimilation (DCA), the water from bathometers was dispensed into 310-mL glass vials using a silicon rubber tube, without turbulence, with double washing, and with overflow of half the vial volume. All vials were darkened during the sample collection. At each horizon, two transparent vials were used for PP measurements, and two darkened ones (covered with aluminum foil) were used for DCA measurements. For DCA control, one darkened vial was fixed with 1 mL 0.05 N HCl. After sampling, 100 µL sterile NaH14CO3 (specific activity 2.04 GBq mmol–1, 5 µCi per sample) was added in the dark. The common darkening sheaths were then removed, and individual sheaths calibrated according to the amount of photosynthetically active radiation at the sampling horizon were applied. The vials were incubated on deck in a flow aquarium for 4 to 6 h. After incubation, the samples were fixed with 1 mL 0.05 N HCl. Subsequent treatment for determination of the PP and DCA rates was carried out as described previously (Savvichev et al., 2020).
The rate of hydrogenotrophic methanogenesis was measured after incubation similar to that for DCA determination and fixation with KOH. After incubation with labeled bicarbonate, the samples were processed as described previously (Pimenov and Bonch-Osmolovskaya, 2006).
To determine the rate of methane oxidation (MO), the water was dispensed into 30-mL penicillin vials with washing and overflow and sealed with rubber stoppers, avoiding air bubbles. After addition of 14CH4 (1 µCi per sample, specific activity 1.16 GBq mmol−1) as solution in degassed distilled water, the samples were incubated for 48 h in a refrigerator under conditions close to in situ ones. Each sample was incubated in two replicates with the control (fixed with 1 N KOH). After incubation the samples were fixed and transported to the laboratory for determination of 14C incorporation into the products of microbial oxidation: СО2, biomass (BM), and dissolved organic carbon (DOC). Sample treatment and calculation of methane oxidation rates were carried as described previously (Rusanov et al., 1998).
Determination of microbial community composition by high-throughput 16S rRNA gene sequencing. The biomass from water samples (1 L) was concentrated on Microsart CN-Filter membranes (Sartorius Stedim Biotech, Germany) with 0.2 µm pore diameter. Prior to DNA isolation in the laboratory the filters were stored in a frozen condition.
DNA was isolated from the biomass using the commercial PowerSoil DNA Isolation Kit (Qiagen, The Netherlands). The 16S rRNA gene libraries were obtained by two sequential PCRs. Both reactions were carried out using the qPCRmix-HS SYBR mixture (Evrogen, Russia) on a StepOnePlus Real-Time PCR System amplifier (Applied Biosystems, United States). During the first reaction, the V4 region of the 16S rRNA genes was amplified using the primers 515F (5′-GTGBCAGCMGCCGCGGTAA-3′) (Hugerth et al., 2014) and Pro-mod-805R (5'-GACTACNVGGGTMTCTAATCC-3′) (Merkel et al., 2019) at the final concentration of 0.25 µM according to the following program: initial denaturing, 3 min at 95°C; 32 cycles (denaturing, 25 s at 95°C; primer annealing, 20 s at 56°C; elongation, 30 s at 72°C); and final elongation, 20 min at 72°C. The amplicons were then used as templates for the second reaction with unique Illumina index primers (TruSeq). The final concentration of the index primers was 0.5 µM. The second PCR was carried out according to the following program: initial denaturing, 3 min at 95°C; 14 cycles (denaturing, 20 s at 95°C; primer annealing, 20 s at 59°C; elongation, 30 s at 72°C); and final elongation, 20 min at 72°C. The libraries were cleaned with the Cleanup Standard kit (Evrogen, Russia), mixed in equimolar amounts, and loaded on 2% agarose gel; the required products were excised on The Safe Imager Blue-Light transilluminator (Invitrogen, Israel) at 470 nm. DNA from the gel was purified using the Cleanup Standard kit (Evrogen, Russia). The final concentrations of the library pools were measured on a Qubit 2.0 fluorimeter with the highly sensitive HS Assay Kit (Life Technologies, United States). Sequencing was carried out on a MiSeq platform (Illumina, United States) using the MiSeq Reagent Micro Kit v2 (300 cycles) according to the manufacturer’s recommendations. Initial processing of the data and removal of chimera sequences was carried out as described previously (Fadrosh et al., 2014). Construction of the OTU tables and alpha-diversity analysis were carried out as described previously (Pimenov et al., 2018). The sequence arrays were deposited to GenBank under BioProject no. PRJNA684336.
RESULTS
Physicochemical characteristics of the stations. Distribution of the hydrophysical and certain hydrochemical parameters in the water column at station AMK73-6045 located above a methane seep, and at the remote station AMK73-6053 is shown on Fig. 2 and in Table 1. The pycnocline was located at 23–32 m at the station above the seep (AMK73-6045) and somewhat higher (20–25 m) at the remote station AMK73-6053. At both stations, the layers above and below the pycnocline were well-aerated, with oxygen concentrations of 340–360 and 320–340 µМ, respectively (Table 1). The concentrations of biogenic elements (P, N) changed insignificantly along the profile. However, calculation of the quasiconservative indicator N*, which reflects variations in the Redfield ratio, revealed nitrogen limitation throughout almost all the water column above the methane seep (AMK73-6045), while its values for the reference station (AMK73-6053) were above zero for all horizons except for the topmost one (Table 1).
Methane concentrations in the water columns of both stations were usually within the range from 25.9 to 50.4 nM. However, at the pycnocline depth (25 m) local maxima were recorded: 61.6 and 116.5 nM for stations AMK73-6045 (seep) and AMK73-6053 (remote), respectively. Methane concentrations in the near-bottom horizons of station AMK73-6045 (seep) was as high as 867.9 nM, while at the reference station it did not exceed the average values until the very bottom (Fig. 2).
Phytoplankton primary production. PP rates revealed at the studied stations were low (Figs. 3a, 3b). The total PP values at stations AMK73-6045 (seep) and AMK-6053 (remote) varied from 2.2 to 18.2 and from 5.2 to 9.9 µg C/(L day), respectively. Integral PP values for two stations were almost the same (Table 2). At different horizons of station AMK73-6045 (seep), the share of bicarbonate carbon (С-НСО3) incorporated in BM varied from 11 to 76%, while the share of С-НСО3 incorporated into DOC varied from 24 to 89% of the summary PP values (Fig. 3a). At the remote station AMK73-6053, carbon incorporation into BM was higher (40 to 75%) and incorporation into DOC was somewhat lower (25 to 60%) (Fig. 3b). On average the PP-associated shares of С-НСО3 incorporation into DOC were 71.8 and 42.7% for stations AMK73-6045 and AMK73-6053, respectively (Table 2).
Microbial processes at stations AMK73-6045 (seep) and AMK73-6053 (remote station) sampled during the 73th cruise of RV Akademik Mstislav Keldysh: primary production (PP, (a–b)), dark CO2 assimilation (DCA, (c–d)), and methane oxidation (MO, (e–f)). Colored bars indicate the ratios of PP, DCA, and MO products during microbial transformation of C-HCO3 (a–d) and C-CH4 (e–f): blue, incorporation into the biomass (BM); red, production of dissolved organic carbon (DOC); and green, СО2 production. The graphs reflect the distribution of the rates of the processes along the profile: (▲) PP, total rate of primary production (BM + DOC); (⚫) DCA, total rate of dark CO2 assimilation (BM + DOC); and (◼) MO, total rate of methane oxidation (СО2 + BM + DOC).
Dark CO2 assimilation (DCA). DCA values measured at two stations were relatively high for Arctic seas during late autumn. At station AMK73-6045 (seep) the total DCA rate varied along the horizons from 2.5 to 4.9 µg C/(L day), with the average value of 3.4 µg C/(L day). The values above the average were found at the surface, in the upper pycnocline (25 m), and in two near-bottom horizons at 65 and 70 m (Fig. 3c). The share of С-НСО3 incorporated into microbial BM varied from 21 to 43% at different horizons, while most С-НСО3 was incorporated to DOC (57−79% of the total DCA).
The DCA depth profile at the remote station AMK73-6053 differed significantly from the one above the seep (Fig. 3d): DCA was above the average in the upper horizons (from the surface to the upper pycnocline) and low from the lower part of the pycnocline to the bottom. However, the DCA range (from 2 to 5.4 µg C/(L day)) and average value (3.8 µg C/(L day)) at the remote station AMK73-6053 were similar to those of station AMK73-6045 above the methane seep. The shares of С-НСО3 incorporation into BM and DOC at different horizons were 17 to 55 and 45 to 83%, respectively.
The average DCA-related shares of С-НСО3 incorporation into BM and DOC were similar at both stations: ~30% into BM and 70% into DOC (Table 2).
Methanogenesis and methane oxidation in the water column. The calculated rate of hydrogenotrophic methanogenesis (MG) determined by radiotracer analysis with NaH14СO3 was quite low, not exceeding 3 nL СН4/(L day), i.e., below the sensitivity of the method, which provides for reliable measurement of MG rates above 5 nL CH4/(L day) at methane concentrations up to 2 µL/L. These results indicate that the contribution of hydrogenotrophic MG to an increase in subsurface methane concentrations in the studied water samples was low or absent.
Microbial methane oxidation (MO) occurred throughout the water column, and methane carbon was converted to СО2, BM, and DOC (Figs. 3e, 3f). MO at both stations were low (except for the near-bottom horizons at the seep station), which is typical of the water column of the open Arctic seas (Rogener et al., 2020). At the remote station AMK73-6053 the range of total methane oxidation rates varied from 9 to 47 nL СН4/(L day), with the average of 20 nL СН4/(L day) (Fig. 3f). Above (15 m) and below the pycnocline (25 m), the values were above the average. The lowest MO rates were detected in the near-bottom horizons (45 and 65 m). Both maxima and minima (the intermediate and near-bottom ones) were in complete agreement with the methane profile (Fig. 2). The shares of MO-related methane carbon incorporation into СО2, BM, and DOC at the reference station were 42.2, 7.9, and 49.9%, respectively (Table 2).
The patterns at station AMK73-6045 above the methane seep were different (Fig. 3e). According to the concentrations of dissolved methane, two zones could be discerned in the water column: from the surface to the lower pycnocline (0–32 m) and from the lower pycnocline to the bottom (Fig. 2). In the zone from the surface to the lower pycnocline, methane concentrations were even lower than at the reference station. The range of MO rates varied uniformly and more significantly: from 16 at the surface to 81 nL СН4/(L day) below the pycnocline, with the average value of 42 nL СН4/(L day) (Fig. 3e). Similar to the reference station, MO rate decreased slightly in the pycnocline, while a local maximum was observed immediately above the pycnocline. Integral MO calculated for this zone was comparable to integral MO for the remote station (Table 2). A drastic increase in methane concentration was, however, revealed at the seep station in the zone from below the pycnocline to the bottom (Fig. 2), as well as a significant (several times) increase in microbial MO rates. The range of methane oxidation increased from 81 (at 32 m) to 120 (at 48 m) and to almost 370 nL СН4/(L day) in the near-bottom horizon (Fig. 3e), while integral MO was several times higher than at the reference station (Table 2).
The distribution of MO-related methane carbon (C-CH4) incorporation into СО2, BM, and DOC calculated for the zone from the surface to the lower pycnocline was different from that calculated for all the water column from surface to bottom (Table 2), indicating the differences in activity of methane-oxidizing microbial communities in the near-bottom and subsurface horizons of the station above the methane seep (AMK73-6045).
Molecular analysis of the composition of microbial communities. Since a peak of methane concentration was revealed in the pycnocline horizon of both stations, three samples were chosen for analysis of microbial phylogenetic diversity, two from the pycnocline of both stations (AMK73-6045_25 and AMK73-6053_25) and one from the horizon above the pycnocline of the seep station (AMK73-6045_16). After primary data processing, the sequencing resulted in 640 to 833 16S rRNA gene sequences with the average length of 254 bp. The reads were combined into operational taxonomic units (OTUs) at 97% similarity. The phenotype coverage calculated by Chao1 varied from 82.8 to 83.8% (Table 3). While the rarefaction curves did not reach the plateau, they were close to this state (supplementary material Fig. S1). While this coverage is insufficient for conclusion on quantitative representation of the minor prokaryotic groups, it is sufficient for description of the dominant taxa.
At the level of higher taxa the composition of three communities was similar. Sequences of the Bacteria domain were more numerous than those of Archaea (over 97% of the total read number) (Fig. 3, Table S1). Archaeal sequences belonged to two phyla: Thaumarchaeota (“Candidatus Nitrosopumilus”; 0.3–1.3%) and Euryarchaeota (Marine Group II; 0.3–0.6%). Most of bacterial sequences belonged to the phylum Proteobacteria (75.6–81.5%), specifically to the classes Alphaproteobacteria (16.5–3.5%), Gammaproteobacteria (42.0–59.3%), and Deltaproteobacteria (0–2.8%). Apart from proteobacteria, members of the phyla Bacteroidetes (9.9–11.5%), Actinobacteria (0.5–1.8%), and Verrucomicrobia (0.6–2.2%) were also revealed. Other phyla of the Bacteria were responsible for not more than 0.9% of the total reads number. Members of the following phylogenetic groups were most numerous in the samples (over 10% of the total reads number): clade SAR11 (class Alphaproteobacteria; 7.2–15.2%), clusters SUP05 (family Thioglobaceae; 7.5–17.4%) and OM60(NOR5) (9.6–10.5%), genus Methyloprofundus (0.1–11.6%), and unidentified members of the family Nitrincolaceae (8.9–13.2%). Members of the genus Planktomarina (2.7–6.4%) and clades SAR92 (4.5–6.6%) and NS5 (2.1–4.3%) were also significant components of the communities. While cyanobacteria were not revealed in the samples, chloroplast sequences were detected (1.3–4.2%). The share of mitochondrial sequences (Rickettsiales) was insignificant (0.3–1.0%).
Among members of the clade SAR11, predominant ones belonged to subclade Ia (6.1–13.4% of the total reads number), which are common in high-latitude areas (Brown et al., 2012). Gammaproteobacterial sequences belonging to the SUP05 cluster were related to the Arctic96BD-19 subcluster, comprising aerobic lithoheterotrophic “Ca. Thioglobus sp.” NP1 and “Ca. Thioglobus singularis” PS1. The OTU sequences from the samples, which were assigned to the cluster OM60(NOR5), were related to the heterotrophic species Marimicrobium arenosum CAU 1038. Unidentified members of the family Nitrincolaceae were related to uncultured clones of marine gammaproteobacteria Arctic96B-16 and Ant4D3.
DISCUSSION
It was earlier found for the central part of the Arctic Ocean that the subsurface methane concentrations were close to the values in equilibrium with the atmosphere (3.5−4 nM) in the Atlantic-derived waters, while it reached 5−7 nM in Pacific-derived waters, which in the second case was associated with the aerobic biological production of methane by pelagic communities (Damm et al., 2010). Methane concentrations in the upper horizons of the water column at the studied stations above the methane seep (AMK73-6045) and at a distance from it (AMK73-6053) had comparable values and were in the range of 25.9−50.4 nM (Table 1), which indicates a significant oversaturation of surface waters of the studied region with methane. Taking the values measured by Damm et al. (2010) as background ones (that is, clearly not determined by the activity of methane seeps), it can be concluded that both studied stations were affected by methane seeps. Thus, the elevated concentration of methane throughout the water column at the AMK73-6045 station was directly caused by the methane bubbles discharge from the bottom sediments, and at the remote station AMK73-6053 – by advective transfer of methane-enriched waters, which can be traced in the pycnocline-associated layer over distances of hundreds of kilometers from a powerful source of bubbly methane (Shakhova et al., 2015). At the studied stations, at the depth of the pycnocline, local maxima of the methane were indeed recorded with the highest value of 116 nM at the station AMK73-6053 (Fig. 2). The highest number (11.6%) of 16S rRNA gene sequences belonging to the methanotrophic bacterial genus Methyloprofundus was also revealed there (sample AMK73-6053_25). Their relative abundance was below 1% in two samples from the station above the seep (AMK73-6045_16 and AMK73-6045_25), which correlates with lower concentrations of dissolved methane (47.3 and 61.6 nM, respectively). The MO rate was also low at both stations, i.e., the contribution of methanotrophic bacteria to the trophic network in the pelagic zone was rather low, unlike the communities of the bottom sediments (Pimenov et al., 2000; Savvichev et al., 2018).
Despite the obvious influence of powerful methane seeps on the concentration of dissolved methane in the water column of the studied region, the possibility and intensity of in situ biological methane production remains a relevant issue. Radiotracer analysis did not reveal measurable hydrogenotrophic methanogenesis in any of the studied water horizons. Methylotrophic MG, which, according to some data, may be a significant source of methane in the oxic water column of the Arctic Ocean (Damm et al., 2010), was not investigated in the present work. However, since no 16S rRNA sequences of methanogenic archaea were revealed in microbial communities of the studied samples (AMK73-6045_16, AMK73-6045_25, and AMK73-6053_25), the possibility of another potential methane producer in the water column should be considered.
The structure and functioning of microbial communities under oligotrophic conditions of Arctic seas are determined by such physicochemical factors as light and availability of biogenic elements and organic matter. According to our data, on average 43 to 75% (and up to 90% at some horizons of the studied stations) of C-CO2 and C-CH4 is converted to DOC in the course of PP, DCA, and MO (Table 2, Fig. 3), which provides for the metabolic activity of diverse groups of pelagic heterotrophic microorganisms. Ability to use, apart from the energy obtained by dissolved organic matter (DOM) decomposition, also the energy of sunlight provides a physiological advantage to photoheterotrophs and proteorhodopsin-containing chemoheterotrophs (Boeuf et al., 2014). Such organisms include aerobic anoxygenic bacteria of the order Rhodobacterales (Amylibacter, Planktomarina, and other Rhodobacteraceae) and proteorhodopsin-containing members of the clades SAR11 (Pelagibacterales), SAR86, and SAR92, which were numerous in the studied samples (Fig. 4, Table S1).
Diversity and relative abundance of prokaryotic phyla (a) and genus-level phylogenetic groups (b) in water samples from stations AMK73-6045 (methane seep) and AMK73-6053 (remote station). Only phylogenetic groups with relative abundance above 1% of the total read number in at least one sample are listed. The overall abundance of phylogenetic groups responsible for less than 1% each is indicated by gray color. The samples AMK73-6045_25 and AMK73-6053_25 were collected from the pycnocline horizon; the sample AMK73-6045_16 was collected from the horizon above the pycnocline. n/i indicates unidentified members of a group.
Ability of the pelagic aerobic community to produce methane is determined by DOM composition and availability of the biogenic elements. Methylphosphonic acid (methylphosphonate, MPn) and dimethylsulfoniopropionate (DMSP) are presently considered the possible methane precursors in oxic marine environments (Karl et al., 2008; Damm et al., 2010). While direct determination of these compounds in our water samples was not carried out, the microorganisms known to be capable of their synthesis and degradation were revealed, and hydrochemical prerequisites for both pathways of methane formation were established.
The genes of methylphosphonate synthesis are known in widespread ammonium-oxidizing marine archaea Nitrosopumilus maritimus (Thaumarchaeota), as well as in members of the SAR11 clade (Pelagibacterales) (Metcalf et al., 2011; Born et al., 2017). Relative abundance of the sequences related to Nitrosopumilus in the studied three samples was low (0.3–1.3% of the total reads number). However, sequences of the SAR11 clade were rather numerous (7.2–15.2%). The latter are able both to synthesize methylphosphonate and to utilize it as a phosphorus source under phosphate limitation, with equimolar amounts of methane released as a byproduct (Carini et al., 2014).
Methylphosphonate demethylation is carried out by CP-lyase. Apart from SAR11, the genes encoding this enzyme were found in various other bacteria and occur widely in the areas of the Global Ocean characterized by phosphorus limitation (Karl et al., 2008; Sosa et al., 2019). In the Arctic region, these conditions occur in the Atlantic-derived waters, including the Laptev Sea. While absolute phosphate concentrations at both stations were not low, positive N* values at the remote station AMK73-6053 indicated an imbalance in the Redfield ratio with nitrogen excess relative to phosphorus, so phosphate starvation at this station may be hypothesized (Table 1). Thus, metabolic activity of both Nitrosopumilus and SAR11, which synthesize and demethylate methylphosphonate, may contribute to aerobic methane production in the studied region.
This suggestion certainly requires experimental verification, since the works on methane release from methylphosphonate via the CP-lyase pathway have been carried out mainly in the low-latitude areas of the Global Ocean (Karl et al., 2008; Sosa et al., 2019), while an opposite pattern has been shown for the central part of the Arctic Ocean, where methane production occurred in the area affected with phosphate-rich Pacific-derived waters, rather than in the zone affected by phosphate-poor Atlantic-derived waters. Thus, methane was released under conditions of phosphate availability and nitrogen limitation. In this case, DMSP was a direct precursor of methane (Damm et al., 2010). Among the studied samples, nitrogen limitation (N* \( \ll \) 0) was observed at station AMK73-6045 above the methane seep (Table 1), where aerobic methane production from DMSP may be expected.
The polar and subpolar seas are known to have elevated DMSP levels, both as a dissolved compound and as a component of microbial biomass. DMSP is synthesized by various members of marine phyto- and bacterioplankton and has a number of functions, including the osmo- and cryoprotector, antioxidant, signal, and regulatory ones, and also acts as a storage of carbon, sulfur, and/or energy (see review by Zhang et al., 2019 and references therein). Among the microorganisms found in our samples, members of the clades SAR11 and SAR86 and of the genus Planktomarina are capable of DMSP utilization (Voget et al., 2015; Sun et al., 2016). The SAR11 cells are unable to assimilate sulfate and thus require such organic sulfur compounds as DMSP, methionine, and cysteine (Tripp et al., 2008). In the Northern Atlantic surface water horizons, the SAR11 cells were found to be responsible for 50% of amino acid assimilation activity and for 30% of DMSP assimilation activity (Malmstrom et al., 2004). Thus, owing to its abundant representation in the community (7.2 to 15.2% of the total read number) and its expected high activity, the clade SAR11 may be the functionally most important component.
DMSP is catabolized via two competing pathways: demethylation with production of methanethiol (methyl mercaptan, MeSH) and cleavage with formation of dimethyl sulfide (DMS) (Zhang et al., 2019). Since both methanethiol and DMS may act as substrates for methylotrophic methanogenesis under anoxic conditions (Tallant and Krycki, 1997), Damm et al. (2015) proposed the theoretical ability of methane production by marine microorganisms in oxic environments. According to their model, calculated for a nominal Roseobacter cell, high respiratory activity and low permeability of the cellular membranes may result in the preservation of anoxic intracellular conditions favoring methane production even in oxygen-saturated waters. A similar mechanism, protecting nitrogenase from oxygen inhibition, is known for nitrogen-fixing microorganisms, including cyanobacteria (Peschek et al., 1991). However, methane production via methylotrophic methanogenesis requires specific enzyme systems (Kurth et al., 2020), which are not known in non-methanogenic prokaryotes. Interestingly, in model experiments elevated numbers of the nifH gene copies were revealed in the Arctic water samples supplemented with DMSP, from which methane was released (Damm et al., 2010). It can be assumed, that methane release from DMSP under nitrogen limitation and nitrogen fixation may be related processes.
To conclude, it should be noted that the pelagic microbial community of the Arctic Seas is an important component of marine trophic networks and may have a significant effect on release of climatically important gases into the atmosphere. High concentration of dissolved methane in the surface, oxygen-rich water of marine basins may result both from discharge of the underwater methane seeps and from activity of subsurface microorganisms in the water column. Our work revealed the hydrochemical prerequisites for and the possible microbial agents of aerobic methane production via methylphosphonate demethylation and DMSP decomposition. Thus, in situ methane production under oxic conditions of the Laptev Sea water column may use DOM, which, according to our data, is extensively synthesized in the processes of PP, DCA, and MO.
Radiotracer techniques, which are traditionally used for measurement of methanogenesis rates, are not suitable for assessment of methane-producing activity of aerobic pelagic microbial communities, since the pathways of methane production by pelagic microorganisms are diverse and are often not associated with utilization of such classical substrates as CO2, acetate, methanol, and methylamines. Further research is therefore required for experimental confirmation of various mechanisms for aerobic methane production by a broad spectrum of heterotrophic microorganisms forming stable communities in the water column of marine basins in various climatic zones. Due to occurrence of both western “heterotrophic” and eastern “autotrophic” provinces (Semiletov et al., 2005; Semiletov et al., 2016), which makes it possible to carry out research in areas with nitrogen and/or phosphorus deficiency during a single cruise, Eastern Arctic seas are attractive subjects for such studies.
Change history
03 August 2021
An Erratum to this paper has been published: https://doi.org/10.1134/S0026261721110011
REFERENCES
Berchet, A., Bousquet, P., Pison, I., Locatelli, R., Chevallier, F., Paris, J.-D., Dlugokencky, E. J., Laurila, T., Hatakka, J., Viisanen, Y., Worthy, D.E.J., Nisbet, E., Fisher, R., France, J., Lowry, D., et al., Atmospheric constraints on the methane emissions from the East Siberian Shelf, Atmos. Chem. Phys., 2016, vol. 16, pp. 4147–4157.
Bianchi, M., Marty, D., Teyssie, J.L., and Fowler, S.W., Strictly aerobic and anaerobic bacteria associated with sinking particulate matter and zooplankton fecal pellets, Marine Ecol. Progr. Ser., 1992, vol. 88, pp. 55–60.
Bižic, M., Grossart, H.-P., and Ionescu, D., Methane paradox, in eLS, Chichester: Wiley, 2020. https://doi.org/10.1002/9780470015902.a0028892
Boeuf, D., Humily, F., and Jeanthon, C., Diversity of Arctic pelagic bacteria with an emphasis on photoheterotrophs: a review, Biogeosciences, 2014, vol. 11, pp. 3309–3322.
Born D.A., Ulrich, E.C., Ju, K.-S., Peck, S.C., van der Donk, W.A., and Drennan, C.L., Structural basis for methylphosphonate biosynthesis, Science, 2017, vol. 358, no. 6368, pp. 1336–1339.
Brown, M.V., Lauro, F.M., DeMaere, M.Z., Muir, L., Wilkins, D., Thomas, T., Riddle, M.J., Fuhrman, J.A., Andrews-Pfannkoch, C., Hoffman, J.M., McQuaid, J.B., Allen, A., Rintoul, S.R., and Cavicchioli, R., Global biogeography of SAR11 marine bacteria, Mol. Syst. Biol., 2012, vol. 8, p. 595.
Carini, P., White, A.E., Campbell, E.O., and Giovannoni, S.J., Methane production by phosphate-starved SAR11 chemoheterotrophic marine bacteria, Nat. Commun., 2014, vol. 5, p. 4346.
Damm, E., Helmke, E., Thoms, S., Schauer, U., Nöthig, E., Bakker, K., and Kiene, R.P., Methane production in aerobic oligotrophic surface water in the central Arctic Ocean, Biogeosciences, 2010, vol. 7, pp. 1099–1108.
Damm, E., Thoms, S., Beszczynska-Möller, A., Nöthig, E., and Kattner, G., Methane excess production in oxygen-rich polar water and a model of cellular conditions for this paradox, Polar Sci., 2015, vol. 9, pp. 327–334.
Ditchfield, A., Wilson, S., Hart, M., Purdy, K.J., Green, D.H., and Hatton, A.D., Identification of putative methylotrophic and hydrogenotrophic methanogens within sedimenting material and copepod faecal pellets, Aquat. Microb. Ecol., 2012, vol. 67, no. 2, pp. 151–160.
Fadrosh, D.W., Ma, B., Gajer, P., Sengamalay, N., Ott, S., Brotman, R.M., and Ravel, J., An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform, Microbiome, 2014, vol. 2, no. 1, p. 6.
Gruber, N. and Sarmiento, J.L., Global patterns of nitrogen fixation and denitrification, Global Biogeochem. Cycles, 1997, vol. 11, no. 2, pp. 235–266.
Hugerth, L.W., Wefer, H.A., Lundin, S., Jakobsson, H.E., Lindberg, M., Rodin, S., Engstrand, L., and Andersson, A.F., DegePrime, a program for degenerate primer design for broad-taxonomic-range PCR in microbial ecology studies, Appl. Environ. Microbiol., 2014, vol. 80, no. 16, pp. 5116–5123.
Karl, D.M., Beversdorf, L., Björkman, K.M., Church, M.J., Martinez, A., and DeLong, E.F., Aerobic production of methane in the sea, Nat. Geosci., 2008, vol. 1, pp. 473–478.
Kurth, J.M., Op den Camp, H.J.M., and Welte, C.U., Several ways one goal—methanogenesis from unconventional substrates, Appl. Microbiol. Biotechnol., 2020, vol. 104, pp. 6839–6854.
Merkel, A.Yu., Tarnovetskii, I.Yu., Podosokorskaya, O.A., and Toshchakov, S.V., Analysis of 16S rRNA primer systems for profiling of thermophilic microbial communities, Microbiology (Moscow), 2019, vol. 88, no. 6, pp. 671–680.
Metcalf, W.W., Griffin, B.M., Cicchillo, R.M., Gao, J., Janga, S.C., Cooke, H.A., Circello, B.T., Evans, B.S., Martens-Habbena, W., Stahl, D.A., and van der Donk, W.A., Synthesis of methylphosphonic acid by marine microbes: a source for methane in the aerobic ocean, Science, 2012, vol. 337, no. 6098, pp. 1104–1107.
Methods of Seawater Analysis, Grashoff, K., Kremling, K., and Ehrhard, M., Eds., Weinheim: Wiley-VCH, 1999.
Malmstrom, R.R., Kiene, R.P., Cottrell, M.T., and Kirchman, D.L., Contribution of SAR11 bacteria to dissolved dimethylsulfoniopropionate and amino acid uptake in the North Atlantic Ocean, Appl. Environ. Microbiol., 2004, vol. 70, pp. 4129–4135.
Peschek, G.A., Villgrater, K., and Wastyn, M., “Respiratory protection” of the nitrogenase in dinitrogen-fixing cyanobacteria, Plant Soil, 1991, vol. 137, pp. 17–24.
Pimenov, N.V. and Bonch-Osmolovskaya, E.A., In situ activity studies in thermal environments, Methods in Microbiology, 35. Extremophiles, Rainey, F.A. and Oren, A., Eds., Amsterdam: Acad. Press, Elsevier, 2006, pp. 29–53.
Pimenov, N.V., Savvichev, A.S., Rusanov, I.I., Lein, A.Yu., and Ivanov, M.V., Microbiological processes of the carbon and sulfur cycles at cold methane seeps of the North Atlantic, Microbiology (Moscow), 2000, vol. 69, no. 6, pp. 709–721.
Pimenov, N.V., Merkel, A.Y., Samylina, O.S., Kanapatskii, T.A., Tikhonova, E.N., Vlasova, M.A., Tarnovetskii, I.Y., and Malakhova, T.V., Structure of microbial mats in the Mramornaya Bay (Crimea) coastal areas, Microbiology (Moscow), 2018, vol. 87, no. 5, pp. 681–691.
Rusanov, I.I., Savvichev, A.S., Yusupov, S.K., Pimenov, N.V., and Ivanov, M.V., Production of exometabolites in the microbial oxidation of methane in marine ecosystems, Microbiology (Moscow), 1998, vol. 67, no. 5, pp. 590–596.
Rogener, M.K., Sipler, R.E., Hunter, K.S., Bronk, D.A., and Joye, S.B., Pelagic methane oxidation in the northern Chukchi Sea, Limnol. Oceanogr., 2020, vol. 65, pp. 96–110.
Savvichev, A.S., Kadnikov, V.V., Kravchishina, M.D., Galkin, S.V., Novigatskii, A.N., Sigalevich, P.A., Merkel, A.Yu., Ravin, N.V., Pimenov, N.V., and Flint, M.V., Methane as an organic matter source and the trophic basis of a Laptev Sea cold seep microbial community, Geomicrobiol. J., 2018, vol. 35, no. 5, pp. 411–423.
Savvichev, A.S., Kadnikov, V.V., Rusanov, I.I., Beletsky, A.V., Krasnova, E.D., Voronov, D.A., Kallistova, A.Yu., Veslopolova, E.F., Zakharova, E.E., Kokryatskaya, N.M., Losyuk, G.N., Demidenko, N.A., Belyaev, N.A., Sigalevich, P.A., Mardanov, A.V., Ravin, N.V., and Pimenov, N.V., Microbial processes and microbial communities in the water column of the polar meromictic Lake Bol’shie Khruslomeny at the White Sea coast, Front. Microbiol., 2020, vol. 11, p. 1945.
Semiletov, I., Dudarev, O., Luchin, V., Shin, K.-H., and Tanaka N., The East-Siberian Sea as a transition zone between Pacific-derived waters and Artic shelf waters, Geophys. Res. Let., 2005, vol. 32, article L10614/2005GL022490.
Semiletov, I., Pipko, I., Gustafsson, Ö., Anderson, L.G., Sergienko, V., Pugach, S., Dudarev, O., Charkin, A., Gukov, A., Bröder, L., Andersson, A., Spivak, E., and Shakhova, N., Acidification of East Siberian Arctic Shelf waters through addition of freshwater and terrestrial carbon, Nature Geosci., 2016, vol. 9, pp. 361–365.
Semiletov, I.P., Pipko I.I., Shakhova, N.E., Dudarev, O.V., Pugach, S.P., Charkin, A.N., McRoy, C.P., Kosmach, D., and Gustafsson, Ö., Carbon transport by the Lena River from its headwaters to the Arctic Ocean, with emphasis on fluvial input of terrestrial particulate organic carbon vs. carbon transport by coastal erosion, Biogeosci., 2011, vol. 8, pp. 2407–2426.
Shakhova, N., Semiletov, I., Leifer, I., Salyuk, A., Rekant, P., and Kosmach, D., Geochemical and geophysical evidence of methane release over the East Siberian Arctic Shelf, J. Geophys. Res., 2010, vol. 115, C08007.
Shakhova, N., Semiletov, I., Sergienko, V., Lobkovsky, L., Yusupov, V., Salyuk, A., Salomatin, A., Chernykh, D., Kosmach, D., Panteleev, G., Nicolsky, D., Samarkin, V., Joye, S., Charkin, A., Dudarev, O., et al., The East Siberian Arctic Shelf: towards further assessment of permafrost-related methane fluxes and role of sea ice, Phil. Trans. R. Soc. A., 2015, vol. 373, article 20140451.
Sosa, O.A., Repeta, D.J., DeLong, E.F., Ashkezari, M.D., and Karl, D.M., Phosphate-limited ocean regions select for bacterial populations enriched in the carbon–phosphorus lyase pathway for phosphonate degradation, Environ. Microbiol., 2019, vol. 21, no. 7, pp. 2402–2414.
Sun, J., Todd, J.D., Thrash, J.C., Qian, Y., Qian, M.C., Temperton, B., Guo, J., Fowler, T.K., Aldrich, J.T., Nicora, C.D., Lipton, M.S., Smith, R.D., De Leenheer, P., Payne, S.H., Johnston, A.W.B., et al., The abundant marine bacterium Pelagibacter simultaneously catabolizes dimethylsulfoniopropionate to the gases dimethyl sulfide and methanethiol, Nature Microbiol., 2016, vol. 1, p. 16065.
Tallant, T.C. and Krycki, J.A., Methylthiol: coenzyme M methyltransferase from Methanosarcina barkeri, an enzyme of methanogenesis from dimethylsulfide and methylmercatopropionate, J. Bacteriol., 1997, vol. 179, pp. 6902–6911.
Tripp, H.J., Kitner, J.B., Schwalbach, M.S., Dacey, J.W.H., Wilhelm, L.J., and Giovannoni, S.J., SAR11 marine bacteria require exogenous reduced sulphur for growth, Nature, 2008, vol. 452, pp. 741–744.
Voget, S., Wemheuer, B., Brinkhoff, T., Vollmers, J., Dietrich, S., Giebel, H.-A., Beardsley, C., Sardemann, C., Bakenhus, I., Billerbeck, S., Daniel, R., and Simon, M., Adaptation of an abundant Roseobacter RCA organism to pelagic systems revealed by genomic and transcriptomic analyses, The ISME J., 2015, vol. 9, pp. 371–384.
Zhang, X.H., Liu, J., Liu, J., Yang, G., Xue, C., Curson, A.R.J., and Todd, J.D., Biogenic production of DMSP and its degradation to DMS—their roles in the global sulfur cycle, Sci. China Life Sci., 2019, vol. 62, no. 10, pp. 1296−1319. https://doi.org/10.1007/s11427-018-9524-y
ACKNOWLEDGMENTS
The authors express their gratitude to the scientific team and ship crew of the 73th cruise of RV Akademik Mstislav Keldysh for the possibilities they provided and for assistance in the sampling procedures.
Funding
The work was supported by the Russian Science foundation (grant no. 16-14-10201) and by the RF Ministry of Science and Higher Education (state assignment for Research Center of Biotechnology, Russian Academy of Sciences). The expedition on the RV Akademik Mstislav Keldysh was organized within the framework of a grant from the Government of the Russian Federation (no. 14.Z50.31.0012/03/ 19.2014). The content of nitrogen compounds was determined by A.A. Grinko supported by the RF Presidential grant for young Cand. Sci. researches MK-535.2020.5. Determination of other hydrochemical parameters was carried out by E.V. Yakushev supported by the RF Ministry of Science and Higher Education (topic 0149-2019-0003).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
Additional information
Translated by P. Sigalevich
The original online version of this article was revised due to a retrospective Open Access order.
Supplementary Information
Rights and permissions
Open Access. This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Samylina, O.S., Rusanov, I.I., Tarnovetskii, I.Y. et al. On the Possibility of Aerobic Methane Production by Pelagic Microbial Communities of the Laptev Sea. Microbiology 90, 145–157 (2021). https://doi.org/10.1134/S0026261721020119
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026261721020119