Abstract—MAPK and PI3K−Akt−mTOR signaling cascades regulate important cellular processes. There was developed the mathematical model that was used for the analysis of the interactions within these cascades. Stability analysis of the PI3K–Ak–mTOR signaling pathway model showed that the baseline state of the signaling pathway is metastable and signal transduction can be activated by overcritical exogenous stimulation. An explicit expression was obtained for the threshold level of external stimuli. It was shown that triggering events in the dynamics of the signaling pathway are described by the fold catastrophe of the elementary catastrophe theory. A discussion about whether the results obtained are significant for molecular hematology and hemostaseology is presented.






Similar content being viewed by others
REFERENCES
B. Alberts, A. Johnson, J. Lewis, et al., Molecular Biology of the Cell (Garland, New York, 2002).
M. J. Berridge, Cell Signalling Biology (Portland Press, London, 2012). doihttps://doi.org/10.1042/csb0001012
Molecular Oncology, Ed. by M. Bishop and R. A. Weinberg (Mosby, 1996).
N. Dey, P. De, B. Leyland-Jones, PI3K−mTOR in Cancer and Cancer Therapy (Humana Press, New York, 2016).
J. Bertacchini, N. Heidari, L. Mediani, et al., Cell. Mol. Life Sci. 72 (12), 2337 (2015).
M. Compagno, Q. Wang, C. Pighi, et al., Nature 542 (7642), 489 (2017).
Z. Li, M. K. Delaney, K. A. O’Brien, and X. Du, Arterioscler. Thromb. Vasc. Biol. 30 (12), 2341 (2010).
G. F. Guidetti, I. Canobbio, and M. Torti. Adv. Biol. Regul. 59, 36 (2015).
A. Martinez-Lopez, L. Salvador-Rodriguez, T. MonteroVilchez, et al., Curr. Opin. Pediatrics 31 (6), 747 (2019).
https://www.genome.jp/kegg-bin/show_pathway?map= hsa04151&show_description=show.
http://public.ndexbio.org/#/network/4bb7e76c-1e5f-11e8-b939-0ac135e8bacf.
B. Pfeuty and K. Kaneko, Phys. Rev. E 89 (2), 022707 (2014).
M. Yčas, On the Nature of the Living: Mechanisms and Meaning (Mir, Moscow, 1994) [in Russian].
B. O. Palsson, Systems Biology: Properties of Reconstructed Networks (Cambridge Univ. Press, Cambridge, 2006).
B. N. Kholodenko, A. Kiyatkin, F. J. Bruggeman, et al., Proc. Natl. Acad. Sci. U. S. A. 99 (20), 12841 (2002).
Yu. M. Romanovskii, N. V. Stepanova and D. S. Chernavskii, Mathematical Biophysics (Nauka, Moscow, 1975) [in Russian].
G. Yu. Riznichenko, Lectures on Mathematical Models in Biology, 2nd ed. (RKhD, Mscow, 2011) [in Russian].
I. A. YAkutik, L. S. Al’-Radi, B. V. Biderman et al., Gematol. Transfuziol. 63, 112 (2018).
A. M. Lyapunov, The General Problem of the Stability of Motion (USSR Acad. Sci., Moscow, 1948; CRC Press, 1992).
A. Nair, P. Chauhan, B. Saha, and K. F. Kubatzky, Int. J. Mol. Sci. 20 (13), 3292 (2019).
D. S. Shen, Y. J. Yang, X. J. Kong, et al., Eur. J. Pharmacol. 852, 1 (2019).
Z. Chen, T. Li, K. Kareem, et al., Artif. Organs 43 (9), 897 (2019).
T. Spater, I. Muller, H. Eichler, et al., Platelets 29 (3), 277 (2018).
J. M. Gibbins, S. Briddon, A. Shutes, et al., J. Biol. Chem. 273 (51), 34437 (1998).
P. Flevaris, Z. Li, G. Zhang, et al., Blood 113 (4), 893 (2009).
N. Nakahata, Pharmacol. Therap. 118 (1), 18 (2008).
Funding
This work was partially supported by the Russian Science Foundation (project no. 19-11-00260).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
CONFLICT OF INTEREST
Authors declare no conflict of interest.
COMPLIANCE WITH ETHICAL STANDARDS
This work does not describe any studies that used humans or animals as objects.
Additional information
Translated by I. Shipounova
Abbreviations: PI3K, phosphatidylinositol-3 kinase; Akt, proteins of the protein kinase B family encoded by AKT genes; mTOR, mammalian target of rapamycin; MAPK, mitogen-activated protein kinase.
SUPPLEMENT
SUPPLEMENT
I. No external stimulation. An analysis of the system of equations (1)–(3) shows that in the absence of a signal from outside (S ≡ 0), the system has two stationary states. One of them has the form
an the second has the form
It is easy to see that the second solution belongs to the region \(Y_{1}^{0}\) ≥ 0, \(Y_{2}^{0}\) ≥ 0, \(Y_{3}^{0}\) ≥ 0 only if the condition is fulfilled
Then, in the absence of an external stimulus, the considered signal system can formally be in one of two stationary states: either in state (S1)–(S3) or in state (S4)–(S6). In order to answer in which state the system can actually be, it is necessary to analyze both stationary states for stability.
Direct research on the first ”method” of A.M. Lyapunov [19] showed that when condition (S7) is satisfied it allows us to find that the stationary state (S1)–(S3) is asymptotically stable with respect to small deviations: all three Lyapunov exponents are negative. Thus, the solution (S1)–(S3) is a node in the phase space of the considered system of equations (1)–(3). A similar study of the stationary solution (S4)–(S6) shows that, of the three Lyapunov exponents, one is positive and the other two are negative. This means that this solution is a saddle in the phase space, i.e., it is unstable to arbitrarily small disturbances and therefore cannot be physically realized.
In addition, it follows from the analysis of the foregoing that, in the complete absence of external stimulation (case S ≡ 0), the dynamics of the signaling path under consideration is actually determined by a pair of equations for Y2 and Y3:
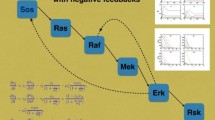
II. Stimulating mTOR with ERK. As can be seen from Fig. 1, the ERK protein is able to directly activate mTOR. This effect can be formally taken into account by adding an additional term to the equation (12). Then, system (10)–(12) takes the following form:
where Λ is value proportional to ERK protein concentration. Given the Λ term, when searching for a stationary state, one of the nullclines will shift down by Λ:
The expression for σ takes the following form:
where the dimensionless parameter is introduced
The solvability condition takes the following form:
Then, the expression for the value of threshold stimulation taking the λ term into account takes the form:
A comparison of expressions (21) and (S17) shows that in the presence of the λ term (λ > 0) the amplitude of the critical level of stimulation σλ is less than σ#. The higher the λ value is, the lower the value of σλ is. Therefore, the MAPK cascade is capable of parametric destabilization of the PI3K–Akt–mTOR signaling pathway.
Rights and permissions
About this article
Cite this article
Sapega, T.S., Guria, G.T. Stability Analysis of the PI3K–Akt–mTOR Signaling Pathway . BIOPHYSICS 65, 259–267 (2020). https://doi.org/10.1134/S0006350920020207
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0006350920020207