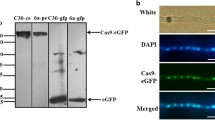
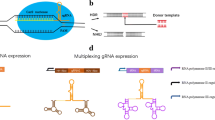
Abstract
The review describes the CRISPR/CAS system and its adaptation for the genome editing in filamentous fungi commonly used for production of enzyme complexes, enzymes, secondary metabolites, and other compounds used in industrial biotechnology and agriculture. In the second part of this review, examples of the CRISPR/CAS technology application for improving properties of the industrial strains of fungi from the Trichoderma, Aspergillus, Penicillium, and other genera are presented. Particular attention is given to the efficiency of genome editing, as well as system optimization for specific industrial producers.




Similar content being viewed by others
Abbreviations
- CRISPR/Cas:
-
clustered regularly interspaced short palindromic repeats and CRISPR-associated protein
- DSB:
-
double-strand break
- HDV:
-
hepatitis D virus
- HH:
-
hammerhead
- HR:
-
homologous recombination
- NHEJ:
-
non-homologous end joining
- NLS:
-
nuclear localization sequence
- sgRNA:
-
single guide RNA
- tracrRNA:
-
trans-activating CRISPR RNA
References
Punt, P. J., van Biezen, N., Conesa, A., Albers, A., Mangnus, J., and van den Hondel, C. (2002) Filamentous fungi as cell factories for heterologous protein production, Trends Biotechnol., 20, 200-206.
Ward, O. P. (2012) Production of recombinant proteins by filamentous fungi, Biotechnol. Adv., 30, 1119-1139, https://doi.org/10.1016/j.biotechadv.2011.09.012.
Lubertozzi, D., and Keasling, J. D. (2008) Developing Aspergillus as a host for heterologous expression, Biotechnol. Adv., 27, 53-57, https://doi.org/10.1016/j.biotechadv.2008.09.001.
Meyer, V. (2008) Genetic engineering of filamentous fungi – progress, obstacles, expression, Biotechnol. Adv., 26, 177-185, https://doi.org/10.1016/j.biotechadv.2007.12.001.
De Vries, R. (2003) Regulation of Aspergillus genes encoding plant cell wall degrading enzymes: relevance for industrial production, Appl. Microbiol. Biotechnol., 61, 10-20, https://doi.org/10.1007/s00253-002-1171-9.
Schuster, A., and Schmoll, M. (2010) Biology and biotechnology of Trichoderma, Appl. Microbiol. Biotechnol., 87, 787-799.
Kumar, R., Singh, S., Singh, O. V. (2008) Bioconversion of lignocellulosic biomass: biochemical and molecular perspectives, J. Industr. Microbiol. Biotechnol., 35, 377-391.
Sinitsyn, A. P., and Rozhkova, A. M. (2015) Penicillium canescens host as the platform for development of new recombinant strains producers of carbohydrases, in Microbiology Monographs, “Microorganisms in Biorefineries” (Kamm, B., ed.) Springer, USA, pp. 1-19.
Sinitsyn, A. P., Sinitsyna, O. A., and Rozhkova, A. M. (2020) Production of industrially important enzyme using Penicillium verruculosum expression system, Biotechnologiya, 36, 17-34.
Harris, D. M, Westerlaken, I., Schipper, D., van der Krogt, Z. A., Gombert, A. K., and Sutherland, J. (2009) Engineering of Penicillium chrysogenum for fermentative production of a novel carbamoylated cephem antibiotic precursor, Metabol. Engin., 11, 125-137.
Ozcengiz, G., and Demain, A. L. (2013) Recent advances in the biosynthesis of penicillins, cephalosporins and clavams and its regulation, Biotechnol. Adv., 31, 287-311.
Corrêa, R. C. G, Rhoden, S. A., Mota, T. R., Azevedo, J. L., Pamphile, J. A., et al. (2014) Endophytic fungi: expanding the arsenal of industrial enzyme producers, J. Ind. Microbiol. Biotechnol., 41, 1467-1478, https://doi.org/10.1007/s10295-014-1496-2.
Toghueo, R. M. K., and Boyom, F. F. (2020) Endophytic Penicillium species nd their agricultural, biotechnological and pharmaceutical application, 3 Biotech, 10, 1-35, https://doi.org/10.1007/s13205-020-2081-1.
Dashtban, M., Schraft, H., and Qin, W. (2009) Fungal bioconversion of lignocellulosic residues; opportunities and perspectives, Int. J. Biol. Sci., 5, 6, 578-595, https://doi.org/10.7150/ijbs.5.578.
Raveendran, S., Parameswaran, B., Ummalyma, S. B., Abraham, A., Mathew, A. K., et al. (2018) Applications of microbial enzymes in food industry, Food Technol. Biotechnol., 56, 16-30, https://doi.org/10.17113/ftb.56.01.18.5491.
MacKenzie, D. A., Jeenes, D. J., and Archer, D. B. (2004) Filamentous Fungi as Expression Systems for Heterologous Proteins, Genetics and Biotechnology (2nd Edn.) Springer-Verlag Berlin-Heidelberg, pp. 289-315.
Kluge, J., Terfehr, D., and Kuck, U. (2018) Inducible promoters and functional genomic approaches for genetic engineering of filamentous fungi, Appl. Microbiol. Biotechnol., 102, 6357-6372, https://doi.org/10.1007/s00253-018-9115-1.
Penttilä, M. (1998) Heterologous protein production in Trichoderma, in Trichoderma and Gliocladium (Kubicek, C. P., and Harman, G. E., eds) Taylor and Francis Ltd., London.
Siedenberg, D., Mestric, S., Ganzlin, M., Schmidt, M., Punt, P. J., et al. (1999) GlaA promoter controlled production of a mutant green fluorescent protein (S65T) by recombinant Aspergillus niger during growth on defined medium in batch and fed-batch cultures, Biotechnol. Progr., 15, 43-50, https://doi.org/10.1021/bp980105u.
Bulakhov, A. G., Volkov, P. V., Rozhkova, A. M., Gusakov, A. V., Nemashkalov, V. A., et al. (2017) Using an inducible promoter of a gene encoding Penicillium verruculosum glucoamylase for production of enzyme preparations with enhanced cellulase performance, PLoS One, 12, e0170404, https://doi.org/10.1371/journal.pone.0170404.
Toews, M. W., Warmbold, J., Konzack, S., Rischitor, P., Veith, D., and Vienken, K. (2004) Establishment of mRFP1 as a fluorescent marker in Aspergillus nidulans and construction of expression vectors for high-throughput protein tagging using recombination in vitro (GATEWAY), Curr. Genet., 45, 383-389.
Dotsenko, G. S., Gusakov, A. V., Rozhkova, A. M., Korotkova, O. G., and Sinitsyn, A. P. (2015) Heterologous beta-glucosidase in a fungal cellulase system: comparison of different methods for development of multienzyme cocktails, Process Biochem., 50, 1258-1263, https://doi.org/10.1016/j.procbio.2015.05.008.
Landowski, C. P., Huuskonen, A., Wahl, R., Westerholm-Parvinen, A., Kanerva, A., et al. (2015) Enabling cost biopharmaceuticals: a systematic approach to delete proteases from a well-known protein production host Trichoderma reesei, PLoS One, 10, e0134723.
Gallo, A., Bruno, K. S., Solfrizzo, M., Perrone, G., Mule, G., Visconti, A., and Baker, S. (2012) New insight into the ochratoxin A biosynthetic pathway through deletion of a nonribosomal peptide synthetase gene in Aspergillus carbonarius, Appl. Environ. Microbiol., 78, 8208-8218, https://doi.org/10.1128/AEM.02508-12.
Ling, S. O. S., Storms, R., Zheng, Yu., Rodzi, M. R. M., Mahadi, N. M., et al. (2013) Development of a pyrG mutant of Aspergillus oryzae strain S1 as a host for the production of heterologous proteins, Sci. World J., 2013, 634317, https://doi.org/10.1155/2013/634317.
Tanaka, M., Ichinose, S., Shintani, T., and Gomi, K. (2018) Nuclear export-dependent degradation of the carbon catabolite repressor CreA is regulated by a region located near the C-terminus in Aspergillus oryzae, Mol. Microbiol., 110, 176-190, https://doi.org/10.1111/mmi.14072.
Todd, R., Lockington, R., and Kelly, J. (2000) The Aspergillus nidulans creC gene involved in carbon catabolite repression encodes a WD40 repeat protein, Mol. Genet. Genom., 263, 561-570, https://doi.org/10.1007/s004380051202.
Tamayo-Ramos, J. A., and Orejas, M. (2014) Enhanced glycosyl hydrolase production in Aspergillus nidulans using transcription factor engineering approaches, Biotechnol. Biofuels, 7, 103, https://doi.org/10.1186/1754-6834-7-103.
Tani, S., Katsuyama, Y., Hayashi, T., Suzuki, H., Kato, M., et al. (2001) Characterization of the amyR gene encoding a transcriptional activator for the amylase genes in Aspergillus nidulans, Curr. Genet., 39, 10-15, https://doi.org/10.1007/s002940000175.
Liu, L., Liu, J., Qiu, R. X., Zhu, X. G., Dong, Z. Y., and Tang, G. M. (2003) Improving heterologous gene expression in Aspergillus niger by introducing multiple copies of protein-binding sequence containing CCAAT to the promoter, Lett. Appl. Microbiol., 36, 358-361.
Zou, G., Shi, S., Jiang, Y., van den Brink, J., de Vries, R. P., et al. (2012) Construction of a cellulase hyper-expression system in Trichoderma reesei by promoter and enzyme engineering, Microb. Cell Factories, 11, 21.
Sun, X., Zhang, X., Huang, H., Wang, Y., Tu, T., et al. (2020) Engineering the cbh1 promoter of Trichoderma reesei for enhanced protein production by replacing the binding sites of a transcription repressor ACE1 to those of the activators, J. Agricult. Food Chem., 68, 1337-1346, https://doi.org/10.1021/acs.jafc.9b05452.
Weld, R. J., Plummer, K. M., Carpenter, M. A., and Ridgway, H. J. (2006) Approaches to functional genomics in filamentous fungi, Cell Res., 16, 31-44, https://doi.org/10.1038/sj.cr.7310006.
Krappmann, S., Sasse, C., and Braus, G. H. (2006) Gene targeting in Aspergillus fumigatus by homologous recombination is facilitated in a nonhomologous end-joining-deficient genetic background, Eukaryot. Cell, 5, 212-215.
Meyer, V., Arentshorst, M., El-Ghezal, A., Drews, A. C., Kooistra, R., and van den Hondel, C. A. (2007) Highly efficient gene targeting in the Aspergillus niger kusA mutant, J. Biotechnol., 128, 770-775.
Ninomiya, Y., Suzuki, K., Ishii, C., and Inoue, H. (2004) Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining, Proc. Natl. Acad. Sci. USA, 101, 12248-12253.
Liu, G., Zhang, J., and Bao, J. (2016) Cost evaluation of cellulase enzyme for industrial-scale cellulosic ethanol production based on rigorous Aspen Plus modeling, Bioproc. Biosyst. Engin., 39, 133-140.
Hu, Y., and Zhu, B. (2016) Study on genetic engineering of Acremonium chrysogenum, the cephalosporin C producer, Synth. Syst. Biotechnol., 1, 3143-3149, https://doi.org/10.1016/j.synbio.2016.09.002.
Peterson, R., and Nevalainen, H. (2012) Trichoderma reesei RUT-C30 – thirty years of strain improvement, Microbiology, 158, 58-68, https://doi.org/10.1099/mic.0.054031-0.
Wu, I., and Arnold, F. (2013) Engineered thermostable fungal Cel6A and Cel7A cellobiohydrolases hydrolase cellulase efficiently at elevated temperatures, Biotechnol. Bioengin., 110, 1874-1883, https://doi.org/10.1002/bit.24864.
Bornscheuer, U. T., Hauer, B., Jaeger, K. E., and Schwaneberg, U. (2019) Directed evolution empowered redesign of natural proteins for the sustainable production of chemicals and pharmaceuticals, Angewande Chemie Int. Edn., 58, 36-40.
Markel, U., Essani, K. D., Besirlioglu, V., Schiels, J., Streit, W. R., and Schwaneberg, U. (2020) Advances in ultrahigh-throughput screening for directed enzyme evolution, Chem. Soc. Rev., 49, 233-262.
Zhang, F., Li, J.-X., Champreda, V., Liu, C.-G., Bai, F.-W., and Zhao, X.-Q. (2020) Global reprogramming of gene transcription in Trichoderma reesei by overexpressing an artificial transcription factor for improved cellulase production and identification of Ypr1 as an associated regulator, Front. Bioengin. Biotechnol., 8, 649, https://doi.org/10.3389/fbioe.2020.00649.
Künkel, W., Berger, D., Risch, S., and Wittmann-Bresinsky, B. (1992) Genetic instability of industrial strains of Penicillium chrysogenum, Appl. Microbiol. Biotechnol., 36, 499-502.
Contreras, F., Thiele, M. J., Pramanik, S., Rozhkova, A., Dotsenko, A. S., et al. (2020) KnowVolution of GH5 Cellulase from Penicillium verruculosum to improve thermal stability for biomass degradation, ACS Sustain. Chem. Engin., 8, 12388-12399, https://doi.org/10.1021/acssuschemeng.0c02465.
Contreras, F., Pramanik, S., Rozhkova, A. M., Zorov, I. N., Korotkova, O. G., et al. (2020) Engineering robust cellulases for tailored lignocellulosic degradation cocktails, Int. J. Mol. Sci., 21, 1589, https://doi.org/10.3390/ijms21051589.
Larue, K., Melgar, M., and Martin, V. J. (2016) Directed evolution of a fungal beta-glucosidase in Saccharomyces cerevisiae, Biotechnol. Biofuels, 9, 52.
Hardiman, E., Gibbs, M., Reeves, R., and Bergquist, P. (2010) Directed evolution of a thermophilic beta-glucosidase for cellulosic bioethanol production, Appl. Biochem. Biotechnol., 161, 301-312.
Sontheimer, E. J., and Barrangou, R. (2015) Origins of the CRISPR genome-editing revolution, Hum. Gene Ther., 26, 413-424, https://doi.org/10.1089/hum.2015.091.
Chen, L., Tang, L., Xiang, H., Jin, L., Li, Q., Dong, Y., et al. (2014) Advances in genome editing technology and its promising application in evolutionary and ecological studies, Gigasciences, 3, 24, https://doi.org/10.1186/2047-217X-3-24.
Liu, W., An, Ch., Sgu, X., Meng, X., Yao, Y., et al. (2020) A dual-plasmid CRISPR/Cas System for mycotoxin elimination in polykaryotic industrial fungi, ACS Public., 9, 2087-2095, https://doi.org/10.1021/acssynbio.0c00178.
Tyagi, S., Kumar, R., Das, A., Won, S.-Y., and Shukla, P. (2020) CRISPR-Cas9 system: a genome-editing tool with endless possibilities, J. Biotechnol., 319, 36-53, https://doi.org/10.1016/j.jbiotec.2020.05.008.
Mali, P, Esvelt, K. M., and Church, G. M. (2013) Cas9 as a versatile tool for engineering biology, Nat. Methods, 10, 957-963, https://doi.org/10.1038/nmeth.2694.
Basset, A. R., and Liu, J. L. (2014) CRISPR/Cas9 and genome editing in Drosophila, J. Genet. Genom., 41, 7-19, https://doi.org/10.1016/j.jgg.20013.12.004.
Blackburn, P. R, Campbell, J. M, Clark, K. J., and Ekker, S. C. (2013) The CRISPR system-keeping zebrafish gene targeting fresh, Zebrafish, 10, 116-118, https://doi.org/10.1089/zeb.2013.9999.
Ebina, H., Misawa, N., Kanemura, Y., and Koyanagi, Y. (2013) Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus, Sci. Rep., 3, 2510, https://doi.org/10.1038/srep02510.
Ishino, Y., Shinagawa, H., Makino, K., Amemura, M., and Nakata, A. (1987) Nucleotide sequence of the iap gene, responsible for alkaline phosphatase conversation in Escherichia coli, and identification of the gene product, J. Bacteriol., 169, 5429-5433.
Mojico, F. J., Diez-Villasenor, C., Soria, E., and Juez, G. (2000) Biological significans of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria, Mol. Microbiol., 36, 244-246.
Jansen, R., van Embden, J. D. A., Gaastra, W., and Schlous, L. M. (2002) Identification of genes that are associated with DNA repeats in prokaryotes, Mol. Microbiol., 43, 1565-1575.
Barrangou, R., Fremax, C., Deveau, H., Richards, M., Boyaval, P., et al. (2007) CRISPR provides acquired resistance against viruses in prokaryotes, Science, 315, 1709-1712.
Nødvig, C. S., Nielsen, J. B., Kogle, M. E., and Mortensen, U. H. (2015) A CRISPR-Cas9 system for genetic engineering of filamentous fungi, PLoS One, 10, e0133085, https://doi.org/10.1371/journal.pone.0133085.
DiCarlo, J. E., Norville, J. E., Mali, P., Rios, X., Aach, J., and Church, G. M. (2013) Genome engineering in Saccharomyces cerevisiae using CRISPR/Cas systems, Nucleic Acids Res., 41, 4336-4343, https://doi.org/10.1093/nar/gkt135.
Horwitz, A. A., Walter, J. M., Schubert, M. G., and Kung, S. H. (2015) Efficient multiplexed integration of synergistic alleles and metabolic pathways in yeasts via CRISPR-Cas, Cell Systems, 1, 88-96, https://doi.org/10.1016/j.cels.2015.02.001.
Curran, K. A., Crook, N., Karim, A., Gupta, A., Wagman, A., and Alpe, H. (2014) Design of synthetic yeast promoters via tuning of nucleosome architecture, Nat. Commun., 5, 4002, https://doi.org/10.1038/ncomms5002.
Doudna, J. A., and Charpentier, E. (2014) The new frontier of genome engineering with CRISPR-Cas9, Science, 346, 1077-1087, https://doi.org/10.1126/science.1258096.
Makarova, K. S., Wolf, Y. I., and Koonin, E. V. (2013) Tha basic building blocks and evoluation of CRISPR/CAS systems, Biochem. Soc. Trans., 41, 1392-1400, https://doi.org/10.1042/BST20130038.
Wang, S., Chen, H., Tang, X., Zhang, H., Chen, W., and Chen, Y. Q. (2017) Molecular tools for gene manipulation in filamentous fungi, Appl. Microbiol. Biotechnol., 101, 8063-8075, https://doi.org/10.1007/s00253-017-8486-z.
Ran, F. A., Hsu, P. D., Lin, C. Y., Gootenberg, J. S., Konermann, S., et al. (2013) Double nicking by RNA-guided CRISPR Cas9 for enchanced genome editing specificity, Cell, 154, 1380-1389, https://doi.org/10.1016/j.cell.2013.08.021.
Hsu, P. D., Lander, E. S., and Zhang, F. (2014) Development and applications of CRISPR-Cas9 for genome engineering, Cell, 157, 1262-1278, https://doi.org/10.1016/j.cell.2014.05.010.
Makarova, K. S., Haft, D. H., Barrangou, R., Brouns, S. J., Charpentier, E., et al. (2011) Evolution and classification of the CRISPR/Cas systems, Nat. Rev. Microbiol., 9, 467-477.
Haft, D. H., Selengut, J., Mongodin, E. F., and Nelson, K. E. (2005) A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR-Cas subtypes exist in prokaryotic genomes, PLoS Computat. Biol., 1, e60.
Chylinsky, K., Le, R. A., and Charpentier, E. (2013) The tracrRNAand Cas9 families of type II CRISPR-Cas immunity systems, RNA Biol., 10, 726-737.
Jinek, M., Chulinski, K., Fonfara, I., Hauer, M., Doudna, J., and Charpentier, E. A. (2012) Programmable dual-RNA-guided DNA endonuclease immunity, Science, 337, 816-821.
Kuscu, C., Arslan, S., Singh, R., Thorpe, J., and Adli, M. (2014) Genome wide analysis reveals characteristics of off-target sites bound by Cas9 endonuclease, Nat. Biotechnol., 32, 677-683.
Cong, L., Ran, F. A., Cox, D., Lin, S., Barreto, R., and Habib, N. (2013) Multiplex genome engineering using CRISPR/Cas systems, Science, 339, 197-217.
Mali, P., Yang, L., Esvelt, K. M., Aach, J., Guell, M., and Dicarlo, J. E. (2013) RNA-guided human genome engineering via Cas9, Science, 339, 823-826.
Jiang, F., Zhou, K., Ma, L., Gressel, S., and Doudna, J. A. (2015) A Cas9-guide RNA complex preorganaized for target DNA recognition, Science, 348, 1477-1481.
Huai, C., Li, G., Yao, R., Zhang, Y., Cao, M., et al. (2017) Structural insights into DNA cleavage activation of CRISPR-Cas9 system, Nat. Commun., 8, 1375.
Stenberg, S. H., Redding, S., Jinek, M., Greene, E. C., and Doudna, J. A. (2014) DNA interrogation by the CRISPR RNA-guided endonuclease Cas9, Nature, 507, 62-67.
Rutkauskas, M., Sinkunas, T., Songailiene, I., Tikhomirova, M. S., Siksnys, V., and Seidel, R. (2015) Directional R-loop formation by the CRISPR/Cas surveillance complex Cascade provides efficient off-target site rejection, Cell Rep., 10, 1534-1543.
Palermo, G., Miao, Y., Walker, R. C., Jinek, M., and McCammon, J. A. (2016) Striking plasticity of CRISPR-Cas9 and key role of non-target DNA, as revealed by molecular simulations, ACS Cent. Sci., 2, 756-763.
Sternberg, S. H., LaFrance, B., Kaplan, M., and Doudna, J. A. (2015) Conformational control of DNA target cleavage by CRISPR-Cas, Nature, 527, 110-113.
Nødvig, C. S., Hoof, J. B., Kogle, M. E., Jarczynska, Z. D., Lehmbeck, J., et al. (2018) Efficient oligo nucleotide mediated CRISPR-Cas9 gene editing in Aspergilli, Fungal Genet. Biol., 115, 78-89, https://doi.org/10.1016/j.fgb.2018.01.004.
Wang, Q., Cobine, P. A., and Coleman, J. J. (2018) Efficient genome editing in Fusarium oxysporum based on CRISPR/Cas9 ribonucleoprotein complexes, Fungal Genet. Biol., 117, 21-29, https://doi.org/10.1016/j.fgb.2018.05.003.
Chen, J., Lai, Y., Wang, L., Zhai, S., Zou, G., et al. (2017) CRISPR/Cas9-mediated efficient genome editing via blastospore-based transformation in entomopathogenic fungus Beauveria bassiana, Sci. Rep., 8, 45763, https://doi.org/10.1038/srep45763.
Chen, B. X., Wei, T., Ye, Z. W., Yun, F., Kang, L. Z., et al. (2018) Efficient CRISPR-Cas9 gene disruption system in edible-medicinal mushroom Cordyceps militaris, Front. Microbiol., 9, 1157, https://doi.org/10.3389/fmicb.2018.01157.
Kislitsin, V. Yu., Chulkin, A. M., Sinel’nikov, I. G., Sinitsin, A. P., and Rozhkova, A. M. (2020) Expression of CAS9 complex of the CRISPR/CAS system for the genome editing of the filamentous fungus Penicillium verruculosum, Vestn. Mosk. Univ. Ser. 2, 61, 47-54.
Matsu-Ura, T., Baek, M., Kwon, J., and Hong, C. (2015) Efficient gene editing in Neurospora crassa with CRISPR technology, Fungal Biol. Biotechnol., 2, 1-7, https://doi.org/10.1186/s40694-015-0015-1.
Sarkari, P., Marx, H., Blumhoff, M. L., Mattanovich, D., Sauer, M., and Steiger, M. G. (2017) An efficient tool for metabolic pathway construction and gene integration for Aspergillus niger, Bioresour. Technol., 245, 1327-1333, https://doi.org/10.1016/j.biortech.2017.05.004.
Pohl, C., Kiel, J. A. K. W., Driessen, A. J. M., Bovenberg, R. A. L., and Nygard, Y. (2016) CRISPR/Cas9 based genome editing of Penicillium chrysogenum, ACS Synthet. Biol., 5, 754-764, https://doi.org/10.1021/acssynbio.6b00082.
Zhang, C., Meng, X., Wei, X., and Lu, L. (2016) Highly efficient CRISPR mutagenesis by microhomology-mediated end joining in Aspergillus fumigatus, Fungal Genet. Biol., 86, 47-57, https://doi.org/10.1016/j.fgb.2015.12.007.
Katayama, T., Tanaka, Y., Okabe, T., Nakamura, H., Fujii, W., and Kitamoto, K. (2016) Development of a genome editing technique using the CRISPR/Cas9 system in the industrial filamentous fungus Aspergillus oryzae, Biotechnol. Lett., 38, 637-642, https://doi.org/10.1007/s10529-015-2015-x.
Schuster, M., Schweizer, G., and Kahmann, R. (2018) Comparative analyses of secreted proteins in plant pathogenic smut fungi and related basidiomycetes, Fungal Genet. Biol., 112, 21-30, https://doi.org/10.1016/j.fgb.2016.12.003.
Schuster, M., Schweizer, G., Reissmann, S., and Kahmann, R. (2016) Genome editing in Ustilago maydis using the CRISPR/Cas system, Fungal Genet. Biol., 89, 3-9, https://doi.org/10.1016/j.fgb.2015.09.001.
Deng, H., Gao, R., Liao, X., and Cai, Y. (2017) Characterization of a major facilitator superfamily transporter in Shiraia bambusicola, Res. Microbiol., 168, 664-672, https://doi.org/10.1016/j.resmic.2017.05.002.
Deng, H., Gao, R., Liao, X., and Cai, Y. (2017) Genome editing in Shiraia bambusicola using CRISPR-Cas9 system, J. Biotechnol., 259, 228-234, https://doi.org/10.1016/j.jbiotec.2017.06.1204.
Lee, C. M., Cradick, T. J., and Bao, G. (2016) The Neisseria meningitidis CRISPR/Cas System enables specific genome editing in mammalian cells, Mol. Ther., 24, 645-654.
Zetsche, B., Gootenberg, J. S., Abudayyeh, O. O., Slaymaker, I. M., Makarova, K. S., et al. (2015) Cpf1 is a single RNA-guided endonuclease of class 3 CRISPR/Cas system, Cell, 163, 759-771.
Swiat, M. A., Dashko, S., den Ridder, M., Wijsman, M., van der Oost, J., et al. (2017) FnCpf1: a novel and efficient genome editing tool for Saccaromyces cerevisiae, Nucleic Acid Res., 45, 12585-12598.
Liu, L., Li, X., Ma, J. Z., Li, Y., Wang, L., Wang, J., et al. (2017) The molecular architecture for RNA-Guided RNA cleavage by Cas13a, Cell, 170, 714-726.
Gao, Y., and Zhao, Y. (2014) Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing, J. Integr. Plant Biol., 56, 343-349, https://doi.org/10.1111/jipb.12152.
Liang, Y., Han, Y., Wang, C., Jiang, C., and Xu, J. R. (2018) Targeted deletion of the USTA and UvSLT2 genes efficiently in Ustilaginoidea virens with the CRISPR-Cas9 system, Front. Plant Sci., 9, 699, https://doi.org/10.3389/fpls.2018.00699.
Zheng, X., Zheng, P., Zhang, K., Cairns, T. C., Meyer, V., Sun, J., and Ma, Y. (2018) 5S rRNA promoter for guide RNA expression enabled highly efficient CRISPR/Cas9 genome editing in Aspergillus niger, ACS Synthet. Biol., 8, 1568-1574.
Nissim, L., Perli, S. D., Fridkin, A., Perez-Pinera, P., and Lu, T. K. (2014) Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells, Mol. Cell, 54, 698-710, https://doi.org/10.1016/j.molcel.2014.04.022.
Wang, J., Li, X., Zhao, Y., Li, J., Zhou, Q., and Liu, Z. (2015) Generation of cell-type-specific gene mutations by expressing the sgRNA of the CRISPR system from the RNA polymerase II promoters, Protein Cell, 6, 689-692, https://doi.org/10.1007/s13238-015-0169-x.
Xue, W., Chen, S., Yin, H., Tammela, T., Papagiannakopoulos, T., et al. (2014) CRISPR-mediated direct mutation of cancer genes in the mouse liver, Nature, 514, 380-384, https://doi.org/10.1038/nature13589.
Shi, T.-Q., Liu, G.-N., Ji, R.-Yu., Shi, K., Song, P., et al. (2017) CRISPR/Cas9-based genome editing of filamentous fungi: the state of the art, Appl. Microbiol. Biotechnol., 101, 7435-7443.
Huang, L., Dong, H., Zheng, J., Wang, B., and Pan, L. (2019) Highly efficient single base editing in Aspergillus niger with CRISPR/Cas9 cytidine deaminase fusion, Microbiol. Res., 223, 44-50, https://doi.org/10.1016/j.micres.2019.03.007.
Liu, R., Chen, L., Jiang, Y., Zhou, Z., and Zou, G. (2015) Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system, Cell Discov., 1, 15007, https://doi.org/10.1038/celldisc.2015.7.
Nielsen, M. L., Isbrandt, T., Rasmussen, K. B., Thrane, U., Hoof, J. B., and Larsen, T. O. (2017) Genes linked to production of secondary metabolites in Talaromyces atroroseus revealed using CRISPR-Cas9, PLoS One, 12, e0169712, https://doi.org/10.1371/journal.pone.0169712.
Salazar-Cerezo, S., Kun, R. S., de Vries, R. P., and Garrigues, S. (2020) CRISPR/Cas9 technology enables the development of the filamentous ascomycete fungus Penicillium subrubescens as a new industrial enzyme producer, Enzyme Microb. Technol., 133, 109463, https://doi.org/10.1016/j.enzmictec.2019.109463.
Yamato, T., Handa, A., Arazoe, T., Kuroki, M., Nozaka, A., et al. (2019) Single crossover-mediated targeted nucleotide substitution and knock-in strategies with CRISPR/Cas9 system in the rice blast fungus, Sci. Rep., 9, 1-8, https://doi.org/10.1038/s41598-019-43913-0.
Liu, Q., Gao, R., Li, J., Lin, L., Zhao, J., et al. (2017) Development of a genome-editing CRISPR/Cas9 system in thermophilic fungal Myceliophthora species and its application to hyper-cellulase production strain engineering, Biotechnol. Biofuels, 10, 1-14, https://doi.org/10.1186/s13068-016-0693-9.
Fuller, K. K., Chen, S., Loros, J. J., and Dunlap, J. C. (2015) Development of the CRISPR/Cas9 system for targeted gene disruption in Aspergillus fumigatus, Eukaryot. Cell, 14, 1073-1080, https://doi.org/10.1128/EC.00107-15.
Fang, Y., and Tyler, B. M. (2016) Efficient disruption and replacement of an effector gene in the oomycete Phytophthora sojae using CRISPR/Cas9, Mol. Plant Pathol., 17, 127-139.
Kuivanen, J., Wang, Y.-M. J., and Richard, P. (2016) Engineering Aspergillus niger for galactaric acid production: elimination of galactaric acid catabolism by using RNA sequencing and CRISPR/Cas9, Microb. Cell Fact., 15, 210.
Weyda, I., Yang, L., Vang, J., Ahring, B. K., Lubeck, M., and Lubeck, P. S. (2017) A comparison of Agrobacterium-mediated transformation and protoplast-mediated transformation with CRISPR-Cas9 and bipartite gene targeting substrates, as effective gene targeting tools for Aspergillus carbonarius, J. Microbiol. Methods, 135, 26-34.
Wenderoth, M., Pinecker, C., Voß, B., and Fischer, R. (2017) Establishment of CRISPR/Cas9 in Alternaria alternate, Fungal Genet. Biol., 101, 55-60.
Weber, J., Valiante, V., Nødvig, C. S., Mattern, D. J., Slotkowski, R. A., and Mortensen, U. H. (2017) Functional reconstitution of a fungal natural product gene cluster by advanced genome editing, ACS Synthet. Biol., 6, 62-68, https://doi.org/10.1021/acssynbio.6b00203.
Wu, C., Chen, Y., Qiu, Y., Niu, X., Zhu, N., et al. (2020) A simple approach to mediate genome editing in the filamentous fungus Trichoderma reesei by CRISPR/Cas9-coupled in vivo gRNA transcription, Biotechnol. Lett., 42, 1203-1210, https://doi.org/10.1007/s10529-020-02887-0.
Zhang, L., Zhao, X., Zhang, G., Zhang, J., Wang, X., et al. (2016) Light-inducible genetic engineering and control of non-homologous end-joining in industrial eukaryotic microorganisms: LML 3.0 and OFN 1.0, Sci. Rep., 6, 20761.
Kocak, D. D., Josephs, E. A., Bhandarkar, V., Adkar, Sh., Kwon, J., and Gersbach, Ch. (2019) Increasing the specificity of CRISPR systems with engineered RNA secondary structures, Nat. Biotechnol., 37, 657-666, https://doi.org/10.1038/s41587-019-0095-1.
Cui, Y., Xu, J., Cheng, M., Liao, X., and Peng, S. (2018) Review of CRISPR/Cas9 sgRNA design tools, Interdiscip. Sci. Computational Life Sci., 10, 455-465.
Kuivanen, J., Arvas, M., and Richard, P. (2017) Clustered genes encoding 2-ketol-gulonate reductase and l-idonate 5-dehydrogenase in the novel fungal d-glucuronic acid pathway, Front. Microbiol., 8, 225, https://doi.org/10.3389/fmicb.2017.00225.
Li, J., Zhang, Y., Zhang, Y., Yu, P. L., Pan, H., and Rollins, J. A. (2018) Introduction of large sequence inserts by CRISPR-Cas9 to create pathogenicity mutants in the multinucleate filamentous pathogen Sclerotinia sclerotiorum, MBio, 9, e00567-e00518, https://doi.org/10.1128/mBio.00567-18.
Miao, J., Li, X., Lin, D., Liu, X., and Tyler, B. M. (2018) Oxysterol-binding protein-related protein 2 is not essential for Phytophthora sojae based on CRISPR/Cas9 deletions, Environ. Microbiol. Rep., 10, 293-298, https://doi.org/10.1111/1758-2229.12638.
Rantasalo, A., Vitikainen, M., Paasikallio, T., Jäntti, J., Landowski, C. P., and Mojzita, D. (2019) Novel genetic tools that enable highly pure protein production in Trichoderma reesei, Sci. Rep., 9, 5032, https://doi.org/10.1038/s41598-019-41573-8.
Kuivanen, J., Korja, V., Holmström, S., and Richard, P. (2019) Development of microtiter plate scale CRISPR/Cas9 transformation method for Aspergillus niger based on in vitro assembled ribonucleoprotein complexes, Fungal Biol. Biotechnol., 6, 3, https://doi.org/10.1186/s40694-019-0066-9.
Dong, L., Lin, X., Yu, D., Huang, L., Wang, B., and Pan, L. (2020) High-level expression of highly active and thermostable trehalase from Myceliophthora thermophila in Aspergillus niger by using the CRISPR/Cas9 tool and its application in ethanol fermentation, J. Industr. Microbiol. Biotechnol., 47, 133-144, https://doi.org/10.1007/s10295-019-02252-9.
Manglekar, R. R., and Geng, A. (2020) CRISPR-Cas9-mediated seb1 disruption in Talaromyces pinophilus EMU for its enhanced cellulase production, Enzyme Microb. Technol., 140, 109646, https://doi.org/10.1016/j.enzmictec.2020.109646.
Rojas-Sánchez, U., López-Calleja, A. C., Millán-Chiu, B. E., Fernández, F., Loske, M., Gómez-Lim, M. A. (2020) Enhancing the yield of human erythropoietin in Aspergillus niger by introns and CRISPR-Cas9, Protein Express. Purif., 168, 105570, https://doi.org/10.1016/j.pep.2020.105570.
Gardiner, D. M., and Kazan, K. (2018) Selection is required for efficient Cas9-mediated genome editing in Fusarium graminearum, Fungal Biol., 122, 131-137, https://doi.org/10.1016/j.funbio.2017.11.006.
Liu, R., Chen, L., Jiang, Y., Zou, G., and Zhou, Z. (2017) A novel transcription factor specifically regulates GH11 xylanase genes in Trichoderma reesei, Biotechnol. Biofuels, 10, 194.
Katayama, T., Nakamura, H., Zhang, Y., Pascal, A., Fujii, W., and Maruyama, J. I. (2019) Forced recycling of an AMA1-based genome-editing plasmid allows for efficient multiple gene deletion/integration in the industrial filamentous fungus Aspergillus oryzae, Appl. Environ. Microbiol., 85, e01896-e01818, https://doi.org/10.1128/AEM.01896-18.
Song, R., Zhai, Q., Sun, L., Huang, E., Zhang, Y., et al. (2019) CRISPR/Cas9 genome editing technology in filamentous fungi: progress and perspective, Appl. Microbiol. Biotechnol., 103, 6919-6932.
Chen, C., Liu, J., Dua, C., Pan, Y., and Liu, G. (2020) Improvement of the CRISPR-Cas9 mediated gene disruption and large DNA fragment deletion based on a chimeric promoter in Acremonium chrysogenum, Fungal Genet. Biol., 134, 103279, https://doi.org/10.1016/j.fgb.2019.103279.
Funding
This work was partially supported by the Russian Foundation for Basic Research (project no. 18-29-07070).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare no conflict of interest. This article does not contain description of studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Rozhkova, A.M., Kislitsin, V.Y. CRISPR/Cas Genome Editing in Filamentous Fungi. Biochemistry Moscow 86 (Suppl 1), S120–S139 (2021). https://doi.org/10.1134/S0006297921140091
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0006297921140091