Abstract
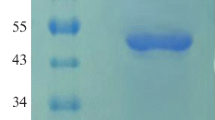
A DNA sequence from Kosakonia sacchari that, according to automated computer analysis, is believed to correspond to the gene for histidine-acid phytase has been selected from the GenBank database. The sequence was optimized for codon composition, synthesized, cloned, and expressed in Pichia pastoris. The main characteristics of the purified recombinant enzyme were determined. It was established that a pH of 4.5 and a temperature of 50°C are optimal for phytase functioning. The specific activity, Michaelis constant (Km), and maximum reaction rate (Vmax) with phytate as a substrate were 1470 U/mg, 193 μM and 2167 μmol/(min mg), respectively. It was shown that the enzyme was characterized by a wide range of working pH levels. Therefore, due to its properties, this new recombinant phytase can be considered a high-potential enzyme for agrobiotechnology.




Similar content being viewed by others
REFERENCES
Yao, M.Z., Zhang, Y.H., Lu, W.L., et al., Phytases: crystal structures, protein engineering and potential biotechnological applications, J. Appl. Microbiol., 2012, vol. 112, pp. 1–14. https://doi.org/10.1111/j.1365-2672.2011.05181.x
Woyengo, T.A. and Nyachoti, C.M., Anti-nutritional effects of phytic acid in diets for pigs and poultry: current knowledge and directions for future research, Can. J. Anim. Sci., 2013, vol. 93, pp. 9–21. https://doi.org/10.4141/cjas2012-017
Rimbach, G., Pallauf, J., and Moehring, J., Effect of dietary phytate and microbial phytase on mineral and trace element bioavailability, Curr. Topics Nutraceut. Res., 2008, vol. 6, no. 3, pp. 131–144.
Dersjant-Li, Y., Awati, A., Schulze, H., and Partridge, G., Phytase in non-ruminant animal nutrition: a critical review on phytase activities in the gastrointestinal tract and influencing factors, J. Sci. Food Agric., 2015, vol. 95, pp. 878–896. https://doi.org/10.1002/jsfa.6998
Humer, E., Schwarz, C., and Schedle, K., Phytate in pig and poultry nutrition, J. Anim. Physiol. Anim. Nutr., 2014, vol. 99, no. 4, pp. 605–625. https://doi.org/10.1111/jpn.12258
Safder, I., Khan, S., Islam, I., and Kazim, M., Pichia pastoris expression system: a potential candidate to express protein in industrial and biopharmaceutical domains, Biomed. Lett., 2018, vol. 4, pp. 1–13.
Maldonado, R.F., Maller, A., Bonneil, E., et al., Biochemical properties of glycosylation and characterization of a histidine acid phosphatase (phytase) expressed in Pichia pastoris,Protein Expr. Purif., 2014, vol. 99, pp. 43–49.
Gessler, N.N., Serdyuk, E.G., Isakova, E.P., and Deryabina, Y.I., Phytases and the prospects for their application (review), Appl. Biochem. Microbiol., 2018, vol. 54, no. 4, pp. 352–360. https://doi.org/10.1134/S0003683818040087
Roy, M.P., Mazumdar, D., Dutta, S., et al., Cloning and expression of phytase appA gene from Shigella sp. CD2 in Pichia pastoris and comparison of properties with recombinant enzyme expressed in E. coli, PLoS One, 2016, vol. 11, e0145745. doi. pone.0145745https://doi.org/10.1371/journal
Bab’eva, I.P. and Golubev, V.I., Metody vydeleniya i identifikatsii drozhzhei (Yeast Isolation and Identification Methods), Moscow: Pishchevaya Promyshlennost’, 1979.
Zhao, X., Huo, K., and Li, Y., Synonymous codon usage in Pichia pastoris,Chin. J. Biotechnol., 2000, vol. 16, pp. 308–311.
Gordeeva, T.L., Borshchevskaya, L.N., Kalinina, A.N., et al., Comparative analysis of the expression efficiency of the bacterial phytase genes in Pichia pastoris yeast by the plate test, Biotekhnologiya, 2017, vol. 33, no. 6, pp. 83–88. https://doi.org/10.21519/0234-2758-2017-33-6-83-88
Gordeeva, T.L., Borschevskaya, L.N., and Sineoky, S.P., Improved PCR-based gene synthesis method and its application to the Citrobacter freundii phytase gene codon modification, J. Microbiol. Methods, 2010, vol. 81, no. 2, pp. 147–152.
Sambrook, J., Fritsch, E.F., and Maniatis, T., Molecular Cloning: A Laboratory Manual, CSHL, 1989, pp. 4–1626.
Chen, C.C., Wu, P.H., Huang, C.T., et al., A Pichia pastoris fermentation strategy for enhancing the heterologous expression of an Escherichia coli phytase, Enzyme Microb. Technol., 2004, vol. 35, no. 4, pp. 315–320. https://doi.org/10.1016/j.enzmictec.2004.05.007
Luo, H., Yao, B., Yuan, T., et al., Overexpression of Escherichia coli phytase with high specific activity, Chin J. Biotechnol., 2004, vol. 20, pp. 78–84.
Gordeeva, T.L., Borshchevskaya, L.N., Kalinina, A.N., et al., Expression and characterization of phytase from Obesumbacterium proteus in the yeast Pichia pastoris, Biotekhnologiya, 2018, vol. no. 4, pp. 18–25. https://doi.org/10.21519/0234-2758-2018-34-4-18-25
Sharp, P.M. and Li, W.H., The codon Adaptation Index—a measure of directional synonymous codon usage bias, and its potential applications, Nucleic Acids Res., 1987, vol. 15, no. 3, pp. 1281–1295.
Teng, D., Fan, Y., Yang, Y., et al., Codon optimization of Bacillus licheniformis β-1,3-1,4-glucanase gene and its expression in Pichia pastoris,Appl. Microbiol. Biotechnol., 2007, vol. 74, pp. 1074–1083. https://doi.org/10.1007/s00253-006-0765-z
Clare, J.J., Rayment, F.B., Ballantine, S.P., et al., High-level expression of tetanus toxin fragment C in Pichia pastoris strains containing multiple tandem integrations of the gene, Biotechnology, 1991, vol. 9, pp. 455–460.
Zhao, W., Xiong, A., Fu, X., et al., High level expression of an acid-stable phytase from Citrobacter freundii in Pichia pastoris,Appl. Biochem. Biotechnol., 2010, vol. 162, pp. 2157–2165. https://doi.org/10.1007/s12010-010-8990-4
Ullah, A.H.J. and Gibson, D.M., Extracellular phytase (E.C. 3.1.3.8) from Aspergillus ficuum NRRL 3135: purification and characterization, Prep. Biochem. Biotechnol., 1987, vol. 17, pp. 63–91.
Golovan, S., Wang, G., Zhang, J., and Forsberg, C.W., Characterization and overproduction of the Escherichia coli appA encoded bifunctional enzyme that exhibits both phytase and acid phosphatase activities, Can. J. Microbiol., 2000, no. 1, pp. 59–71.
Xiong, A.S., Yao, Q.H., Peng, R.H., et al., High level expression of a synthetic gene encoding Peniophora lycii phytase in methylotrophic yeast Pichia pastoris,Appl. Microbiol. Biotechnol., 2006, vol. 72, no. 5, pp. 1039–1047. https://doi.org/10.1007/s00253-006-0384-8
Shi, P., Huang, H., Wang, Y., et al., A novel phytase gene appA from Buttiauxella sp. GC21 isolated from grass carp intestine, Aquaculture, 2008, vol. 275, pp. 70–75.
Tai, H.M., Yin, L.J., Chen, W.C., and Jiang, S.T., Overexpression of Escherichia coli phytase in Pichia pastoris and its biochemical properties, J. Agric. Food Chem., 2013, no. 25, pp. 6007–6015.
Kim, H.W., Kim, Y.O., Lee, J.H., et al., Isolation and characterization of a phytase with improved properties from Citrobacter braakii,Biotechnol. Lett., 2003, vol. 25, pp. 1231–1234.
Huang, H., Luo, H., Yang, P., et al., A novel phytase with preferable characteristics from Yersinia intermedia,Biochem. Biophys. Res. Commun., 2006, vol. 350, pp. 884–889. https://doi.org/10.1016/j.bbrc.2006.09.118
Liebert, F., Wecke, C., and Schoner, F.J., Phytase activities in different gut contents of chickens are dependent on level of phosphorus and phytase supplementations, in Proceedings of 1st European Symposium Enzymes in Animal Nutrition, Wenk, C. and Boessinger, M., Eds., Karthause Ittingen, Switzerland, 1993, pp. 202–205.
Dersjant-Li, Y., Awati, A., Schulze, H., and Partridge, G., Phytase in non-ruminant animal nutrition: a critical review on phytase activities in the gastrointestinal tract and influencing factors, J. Sci. Food Agric., 2015, vol. 95, no. 5, pp. 878–896. https://doi.org/10.1002/jsfa.6998
Neira-Vielmaa, A.A., Aguilara, C.N., Ilyinab, A., et al., Purification and biochemical characterization of an Aspergillus niger phytase, produced by solid-state fermentation using triticale residues as substrate, Biotechnol. Rep., 2018, vol. 17, pp. 49–54.
Welker, J.S., Rosa, A.P., Moura, D.J., et al., Temperatura corporal de frangos de corte em diferentes sistemas de climatização, Revista Brasileira de Zootecnia, 2008, vol. 3, no. 8, pp. 1463–1467.
Humer, E., Schwarz, C., and Schedle, K., Phytate in pig and poultry nutrition, J. Anim. Physiol. Anim. Nutr., 2015, vol. 99, pp. 605–625.
ACKNOWLEDGEMENTS
The work was carried out at the Unique Scientific Facility of the All-Russia Collection of Industrial Microorganisms National Bioresource Center, Kurchatov Institute NRC, GOSNIIGENETIKA.
Funding
The work was financially supported by the Ministry of Education and Science of Russian Federation (Unique Project Identifier RFMEFI57917X0145).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflicts of interest.
This article does not contain any studies involving animals or humans performed by any of the authors.
Additional information
Translated by I. Gordon
Abbreviations: aa—amino-acid residue(s); CAI—codon adaptation index; CL—culture liquid; PAGE—polyacrylamide gel electrophoresis; PCR—polymerase chain reaction; SDS—sodium dodecyl(lauryl)sulfate.
Rights and permissions
About this article
Cite this article
Gordeeva, T.L., Borshchevskaya, L.N., Kalinina, A.N. et al. New Recombinant Phytase from Kosakoniasacchari: Characteristics and Biotechnological Potential. Appl Biochem Microbiol 56, 779–786 (2020). https://doi.org/10.1134/S0003683820070042
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0003683820070042