Abstract
Stenotrophomonas sp. is emerging as a popular microbe of global concern with various potential ecological roles. Biosynthesis of gold and silver nanoparticles (AgNPs) using this bacterial strain has shown promising applications in life sciences. However, there is no report on efficient agricultural applications of biosynthesized AgNPs using Stenotrophomonas sp. In this regard, successful biosynthesis of AgNPs using Stenotrophomonas sp. BHU-S7 (MTCC 5978) was monitored by Uv-visible spectrum showing surface plasmon resonance (SPR) peak at 440 nm. The biosynthesized AgNPs were spherical with an average mean size of ~12 nm. The antifungal efficacy of biosynthesized AgNPs against foliar and soil-borne phytopathogens was observed. The inhibitory impact of AgNPs (2, 4, 10 μg/ml) on conidial germination was recorded under in vitro conditions. Interestingly, sclerotia of Sclerotium rolfsii exposed to AgNPs failed to germinate on PDA medium and in soil system. Moreover, AgNPs treatment successfully managed collar rot of chickpea caused by S. rolfsii under greenhouse conditions. The reduced sclerotia germination, phenolic acids induction, altered lignification and H2O2 production was observed to be the probable mechanisms providing protection to chickpea against S. rolfsii. Our data revealed that AgNPs treated plants are better equipped to cope with pathogen challenge pointing towards their robust applications in plant disease management.
Similar content being viewed by others
Introduction
The genus Stenotrophomonas, previously named as Pseudomonas maltophilia, phylogenetically belongs to γ-subclass of Proteobacteria1,2. This novel genus was first proposed by Palleroni and Bradbury3 after studying the notable differences between P. maltophilia and closely associated xanthomonads group. Later, on the basis of heterogeneity on the phenotypic and genotypic level, seven species of Stenotrophomonas has been recognized so far between 1993 and 20074. This ubiquitous microorganism is found in a broad range of ecological niches and has been identified as a major soil inhabitant5,6. Stenotrophomonas sp. has gathered considerable focus of scientific community due to its important ecological role in bioremediation7,8, element cycle9, plant growth promotion10,11 and biocontrol of phytopathogens12,13,14,15.
Many species of Stenotrophomonas have versatile distribution and are found to be widely associated with plants. In relation to agriculture, Stenotrophomonas species have the potential to be used as an efficient candidate for plant growth promotion and biocontrol of phytopathogens. Well documented studies have reported that Stenotrophomonas species produce plant growth hormone and also generate other factors directly responsible for plant growth promotion10. In addition, they also produce antibiotics, volatile organic compounds and fungal cell wall degrading enzymes indicating their key contribution to the biocontrol activity against several soil-borne plant pathogens6,16,17. Furthermore, Dunne et al.15 elucidated the biocontrol potential of S. maltophilia W81 isolated from sugarbeet rhizosphere against damping-off disease causing Pythium ultimum. Interestingly, authors also explained production of an extracellular protease as the possible mechanism behind its biocontrol activity. Similarly, in another study Zhang et al.16 reported that chitinase production by S. maltophilia C3 is mainly responsible for its antifungal activity against broad range of phytopathogens. For a long time, S. maltophilia has been recognized as opportunist human pathogen but later on, another non-pathogenic species i.e. S. rhizophila within the complex of S. maltophilia was also identified18,19. S. rhizophila is considered as an important plant growth promoting bacteria and found to be associated with both rhizosphere and phylloplane20. This species is differentiated from S. maltophilia on the basis of presence of glucosyl glycerol osmolyte (found only in S. rhizophila) and hence it exhibits high degree of salt tolerance21. Previous report by Egamberdieva et al.22 demonstrated that S. rhizophila DSM14405T mediated growth promotion of various crops in highly salinated soil of Uzbekistan. Congruently, these exciting studies indicate enormous potential applications of Stenotrophomonas sp. in agriculture owing to their advantageous interactions with plants.
Remarkably, use of such beneficial microbes as biological inoculants for improving plant growth, biological control of plant diseases and amelioration of stressed environment, offers an environment friendly and sustainable approach23. However, their inconsistent performance under field conditions pertaining to several factors limits their practical utility. To overcome such obstacles affecting the translation of biological inoculants into natural environment, the agricultural sector is now exploiting the innovative approach of nanotechnology based strategies24,25,26,27. In context to this, several biological agents including microbes and plants are being utilized for biosynthesis of nanoparticles24,28. Moreover, the biosynthesized nanoparticles are ecofriendly and safe showing robust applications in diverse fields viz. pharmaceuticals, cosmetics, medicine, agriculture etc.29,30. Numerous reports have deciphered successful and efficient applications of biosynthesized nanoparticles for agricultural purposes particularly for plant growth and plant disease management24,26,31,32,33,34,35. These exciting studies and above mentioned potential efficiency of Stenotrophomonas sp. in agriculture distinctly provide a substantial base for harnessing the nanoparticles synthesis property of this strain and its further utility for agricultural benefits36. However, few earlier reports have demonstrated Stenotrophomonas sp. isolated from various ecological niches, as a potential candidate for biosynthesis of gold and silver nanoparticles37,38,39. Moreover, these biofabricated nanoparticles also showed promising antibacterial and cytotoxic effects. Interestingly, it is worth mentioning here that so far there have been no reports on the potential role of biosynthesized nanoparticles using this strain in agriculture primarily for plant growth improvement and plant diseases management. In relation to this, the present study investigates the synthesis of silver nanoparticles (AgNPs) by Stenotrophomonas sp. BHU-S7 (MTCC-5978) isolated from soil samples from agricultural fields and its efficient role as nanofungicide against a broad range of phytopathogenic fungi. Moreover, we also aim to provide a direct evidence of the inhibitory effect of biosynthesized AgNPs against soil-borne phytopathogen Sclerotium rolfsii, causing collar rot of chickpea under greenhouse condition.
Materials and Methods
Isolation and identification of bacterial strain
Bacterial strain BHU-S7 was isolated from soil sample collected from agricultural farm of Banaras Hindu University, Varanasi (25°20′ N latitude & 83°01′ E longitude) using the protocol as described earlier24. Pure culture of BHU S-7 was maintained on nutrient agar (NA) (HI-MEDIA Laboratories, Bombay, India).
16S rDNA sequencing of bacterial strain
The bacterial strain BHU-S7 was identified using 16S rDNA sequence analysis using universal primer pair, forward (5′AGAGTTTGATCCTGGCTCAG3′) and reverse (5′AAGGAGGTGATCCAGCCGCA 3′)40. The analysis of partial 16S rDNA sequence was done using BLAST program at National Center for Biotechnology Information (NCBI). The closely related sequences were selected and aligned using Clustal X and further phylogenetic tree was constructed by neighbor-joining method using MEGA 6.0 software.
AgNPs biosynthesis by bacterial strain BHU-S7 (MTCC 5978)
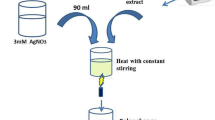
The AgNPs were biosynthesized by bacterial strain BHU-S7 following the protocol as described earlier by Mishra et al.24. Aqueous solution of 1 mM AgNO3 prepared in sterile distilled water was used for AgNPs synthesis purpose. Briefly, culture supernatant obtained from 48 h grown culture of bacterial strain BHU-S7 in Nutrient Broth (NB) media was treated with 1 mM AgNO3 solution and incubated in dark at 30 °C. The biosynthesis of AgNPs was confirmed by visual color change of the solution from yellow to brown. After the completion of synthesis process, AgNPs were collected in the form of pellet by centrifuging the synthesis mixture at 14,000 rpm for 30 min. The obtained pellet was washed several times with sterile distilled water for better removal of any medium components and unconverted Ag+ ions as well. The resulting AgNPs in the form of pellet was freeze dried at −80 °C and used for further studies33.
Characterization of biosynthesized AgNPs
The optical characterization of biosynthesized AgNPs was carried out using UV-visible spectroscopy. The absorption spectra of biosynthesized AgNPs solution was monitored by UV–visible spectrophotometer (Perkin Elmer Life and Analytical Sciences, CT, USA) in the range of 300–600 nm. In order to adjust the baseline, nutrient broth media was used as blank. For structural characteristic, X-ray diffraction (XRD) pattern of biosynthesized nanoparticles was performed as described by Singh et al.41 employing MiniFlex™ II benchtop XRD system (Rigaku Corporation, Tokyo, Japan) operated at 40 kV with Cu-Kα radiation in the range of 2θ = 20°−80°. Further, size and morphology of biosynthesized AgNPs were analysed by scanning electron microscopy (SEM) using JSM 6510LV scanning electron microscope (JEOL, Tokyo, Japan) and transmission electron microscopy (TEM) using JEOL 100/120 kV TEM (JEOL, Tokyo, Japan) operated at accelerating voltage of 20 kV and 200 kV respectively. The Fourier transform infrared (FTIR) spectrum for analyzing functional nature of AgNPs was recorded on Perkin Elmer FT-IR spectrometer Spectrum Two (Perkin Elmer Life and Analytical Sciences, CT, USA) using potassium bromide (KBr) pellets in the ratio of 1:100. The spectrum was recorded in the diffuse reflectance mode at a resolution of 4 cm−1 in the range of 400–4000 cm−1 wavenumber42. For elemental composition, energy dispersive X-ray (EDAX) spectrum was collected using EDAX detector (Oxford Instruments INCAx-sight EDAX spectrometer) equipped with SEM. Furthermore, the thermogravimetric analysis (TGA) was conducted using Pyris1 thermogravimetric analyzer (TGA; Perkin Elmer Life and Analytical Sciences, CT, USA) in order to check thermal stability of biosynthesized AgNPs. For this, sample was subjected to temperature range of 50–700 °C at a heating rate of 10 °C/min under nitrogen atmosphere.
In vitro assay for antifungal activity of biosynthesized AgNPs
The antifungal activity of biosynthesized AgNPs was checked against foliar (Alternaria alternata, Curvularia lunata and Bipolaris sorokiniana) and soil borne (Sclerotium rolfsii) phytopathogens. In case of foliar pathogens, a cavity slide method was used as described by Mishra et al.24. For this, conidial suspension was harvested in sterile distilled water from pure and freshly grown culture of A. alternata, C. lunata and B. sorokiniana on Potato Dextrose Agar (PDA) media under aseptic condition. Three experimental sets i.e. control, AgNO3 control and AgNPs treated sets were prepared in triplicate for each test fungus. In treated set, the conidial suspension was mixed with 2, 4 and 10 μg/ml of AgNPs and filled in cavity slide. On other side, the control set was maintained by filling the cavity with conidial suspension alone while AgNO3 control was maintained by mixing 2, 4 and 10 μg/ml of AgNO3 with conidial suspension. The cavity slides from all sets were kept carefully inside the petriplate containing sterilized moist blotting paper. These moist chambers containing cavity slides were incubated at 25 ± 2 °C for 24 h. The slides were examined under light microscope (Nikon DS-fi1, Japan) to observe the inhibitory effect of AgNPs on conidial germination.
Further, inhibitory effect of biosynthesized AgNPs on sclerotia germination of Sclerotium rolfsii was investigated by dipping the sclerotia in direct suspension of AgNPs for 12 and 24 h. Later sclerotia were kept on PDA plates for their germination. Sclerotia dipped in sterile distilled water served as control. For more accuracy, sclerotia germination was also checked in soil. For this, control and AgNPs treated sets were prepared in triplicate by placing 25 g garden soil in petriplate. The sclerotia was placed into the soil and drenched with AgNPs suspension maintaining 16% moisture while in control set, soil was drenched with water only. Petriplates were kept in BOD incubator at 25 ± 2 °C for a week.
Tripartite interaction among Cicer arietinum, Sclerotium rolfsii and antifungal biosynthesized AgNPs under Greenhouse conditions
The antifungal efficacy of biosynthesized AgNPs was evaluated against collar rot disease caused by S. rolfsii (SR) under greenhouse conditions using chickpea (cv. Avrodhi) as a host plant. For conducting greenhouse experiment, plastic pots (5 inch length, 13 cm diameter) were filled with soil (650 g) and 5 chickpea seeds were sown in each pot. For proper seed germination and plant growth, pots were watered to maintain adequate soil moisture. After 15 days of plant growth, following treatments were maintained: (i) unchallenged control (C), (ii) pathogen challenged control (SR control) and (iii) pathogen challenged + AgNPs (SR + AgNPs). Six replicates for each treatment were maintained. The pathogen (S. rolfsii) was introduced to the plants by placing one sclerotium adjacent to the collar region of chickpea in each replicate and further covered with moist cotton for maintaining the humidity required for sclerotial germination. For SR + AgNPs treatment, after placing sclerotia on the collar region, AgNPs suspension was sprayed with automizer over this and then covered with absorbant cotton soaked in AgNPs suspension. The infection in collar region by S. rolfsii was observed after 24 h of inoculation.
Chromatographic detection and quantification of phenolic compounds in chickpea
Extraction of phenolic compounds was done following the protocol described by Niranjan et al.43. For extraction purpose, 1.0 g of plant sample collected (after 24 h of pathogen challenge) in triplicate from each treatment is further processed separately in order to obtain the data in triplicate. Plant sample was crushed with 10 ml of methanol:water:hydrochloric acid solution (5:4:1, v/v) in a mortar and pestle and left overnight in orbital shaker at room temperature. After this, the mixture was filtered through Whatman No. 1 filter paper and plant residue was further extracted twice with same solution for maximum extraction of phenolic compounds. The filtered plant extract was combined together and concentrated upto half of the volume using rotary evaporator (Eyela N–N series, Tokyo, Japan). The residue was further fractionated three times with Ethyl acetate. The collected ethyl acetate fraction was concentrated under reduced pressure on rotary evaporator. The obtained residue was dissolved in 1.0 ml of HPLC grade methanol and filtered through a 0.2 μm membrane filter (Merck). This stock solution was subjected to HPLC for detection and quantification of phenolic compounds.
For HPLC analysis, 20 μl of sample was injected into C-18 HPLC (4 μm, 250 × 4.6 mm, Phenomenex, USA) using HPLC system (Shimadzu, Japan) equipped with LC-10 dual pump and UV detector SPD-10A. The mobile phase used for compounds separation was a linear gradient prepared from acetic acid (1% v/v) in HPLC grade water (pump A) and acetonitrile (pump B), with detection at 254 nm. The flow rate was maintained at 1.0 ml/min. Results were obtained by comparisons with standards using data integrated by Shimadzu class VP series software. Phenolic compounds were detected in sample by comparing retention time (RT) of peaks with that of standard and further quantified by comparing peak area in the sample with that of standard.
H2O2 detection in leaves by DAB (3,3′-diaminobenzidine) staining method
Subcellular localization of H2O2 in chickpea leaves was done using the protocol described by Thordal-Christensen et al.44. Leaf sample was collected from 6 independent plants of each treatment. DAB staining solution is prepared by dissolving 50 mg of DAB in 45 ml of sterile distilled water and final pH was set to 3 using 0.2 M HCl. After this 0.05% of Tween 20 together with 2.5 ml of 200 mM Na2HPO4 was added to DAB solution. DAB solution is stored in dark brown bottle as it is light sensitive. For staining purpose, leaf sample was dipped overnight in DAB solution in dark condition at room temperature. After this, leaves were removed from DAB solution and further boiled in bleaching solution containing alcohol and acetic acid (3:1) till leaves become clear (completely devoid of chlorophyll). The boiling step will bleach out the chlorophyll so that brown precipitate formed by reaction of DAB with H2O2 remains in the leaves. For detection of brown precipitate, leaves were visualized under light microscope (Nikon DS-fi1, Japan).
Histochemical staining for stem lignification
After 24 h of pathogen challenge, plants were harvested and chickpea stem was used for microscopic investigation of lignification pattern in each treatment using the protocol as described earlier24,45. Transverse sections of stem were stained in saturated aqueous solution of phloroglucinol in 20% HCl and immediately observed under light microscope for lignification as indicated by red-violet color.
Results and Discussion
Identification of BHU-S7
The bacterial strain BHU-S7, isolated from agricultural farm soil was identified as Stenotrophomonas sp. using 16S rDNA sequencing analysis. The nucleotide sequence of 1445 bp was phylogenetically characterized using BLAST tool of NCBI search. The constructed phylogenetic tree indicated that the bacterial strain BHU-S7 shared a maximum 99% close homology with Stenotrophomonas sp. (Fig. 1).
Phylogenetic tree of bacterial strain BHU S-7 constructed based on 16s rDNA sequence. It is identified as Stenotrophomonas sp. BHU S-7 (as indicated by asterisk). The partial 16S rDNA sequence has been deposited in NCBI GenBank under the accession number KF863907. Bootstrap values are based on 1,000 replications. Bar 0.005 substitutions per site.
Nucleotide sequence accession number and MTCC number
The partial nucleotide sequences for 16S rDNA of the bacterial strain BHU S-7 has been submitted to Genbank under the accession number KF863907. Furthermore, pure culture of the bacterial strain has also been deposited at IMTECH, Chandigarh, India with MTCC number 5978.
Biosynthesis of AgNPs by Stenotrophomonas sp. BHU-S7
In this study, Stenotrophomonas sp. BHU-S7 isolated from agricultural soil was used for biosynthesis of AgNPs. The visible appearance of dark brown color in culture supernatant upon addition of AgNO3 confirms successful extracellular synthesis of AgNPs. In the whole biosynthesis process, the noticeable color change of culture supernatant from yellow to dark brown is due to reduction of silver ions Ag+ into Ag° by using active biomolecules present in culture supernatant46,47. However, no color change was observed in control set where plain growth media was treated with 1 mM AgNO3 indicating the functional involvement of bacterial biomolecules in reduction process leading to biosynthesis of AgNPs. The development of brown color in the reaction mixture is mainly attributed to excitation of surface plasmon resonance, a unique optical property of noble metals48,49. Interestingly, the most widely accepted mechanism for extracellular biosynthesis of AgNPs is via extracellular enzymes such as nitrate reductase enzyme present in supernatant that help in transferring the electron to silver ion and hence reduced to AgNPs47,50.
Characterization of biosynthesized AgNPs
Optical
The unique surface plasmon resonance (SPR) property of noble metals strongly induces the absorption of the incident light and thus can be monitored by Uv-Visible spectroscopy51,52. Moreover, this SPR band is more prominent for noble metal nanoparticles particularly for AgNPs and AuNPs53. Hence, in the present study, the biosynthesis of AgNPs was ascertained by Uv-visible spectroscopy that displayed a strong surface plasmon resonance (SPR) peak at 440 nm (Fig. 2). It is evident from previous reports that AgNPs exhibit UV-visible absorption spectra in the range of 400–500 nm due to Mie scattering51. Therefore, presence of characteristic sharp and clear peak at 440 nm indicates successful biosynthesis of AgNPs. While on other side, this distinctive peak was absent in control set signifying no reduction of Ag ions. The reduction process of silver ions and the biosynthesis of stable AgNPs occurred within 24 hour of reaction.
Structural
The crystallinity of biosynthesized AgNPs was confirmed by X-ray diffraction (XRD) analysis that determines size, lattice and structure of nanoparticles54. The diffractogram showed well resolved four diffraction peaks at 2θ angles of 38.39°, 49.25°, 64.10°, and 77.87° which were indexed to the Bragg reflection peaks of (111), (200), (220), and (311) planes of face centered cubic (fcc) structures of AgNPs, respectively (Fig. 3a). In this experiment, the XRD data confirmed that the synthesized nanoparticles were crystalline in nature and had a face-centered cubic structure. Further, the average crystallite size (d) of biosynthesized AgNPs was calculated following the Debye-Scherrer formula:

where k = 0.9 is the shape factor, λ is the X-ray wavelength of Cu Kα radiation (1.54 Å), θ is the Bragg diffraction angle, and B is the full width at half maximum of the (111) plane diffraction peak55. The calculated average crystallite size was estimated to be 12.7 nm.
Morphological
The morphological investigation of the biosynthesized AgNPs was carried out using SEM and TEM analysis. The Fig. 3b shows the SEM micrograph recorded from dry mass of biosynthesized AgNPs, which was deposited on a carbon tape. The micrograph clearly demonstrated the aggregation of micro particles under the powder form. However, the narrow distribution of AgNPs in the TEM analysis was in accordance with the UV-visible spectral study. The presence of single SPR peak as revealed by UV-visible spectroscopy divulges spherical shape of AgNPs56. In the present study, TEM analysis confirms the existence of spherical AgNPs with slight agglomerations at some places that may be due to presence of biological macromolecules on their surfaces (Fig. 3c). Moreover, AgNPs represented variable size ranging from 5 to 30 nm with average mean size estimated to be ~12 nm. However, such variations in size are probably due to the fact that AgNPs were being synthesized at different times57.
Elemental
The compositional analysis of the biosynthesized AgNPs was carried out by Energy dispersive X-ray (EDAX). The EDAX spectra displayed characteristic signal peak of silver element (Ag) that confirmed the crystallinity and mettalic nature of biosynthesized AgNPs (Fig. 4a). It is worth mentioning here that silver nanocrystallites exhibit distinctive absorption peak at 3 keV because of surface plasmon resonance58. Therefore, this data reveal successful synthesis of AgNPs as maximum proportion of total elemental composition is covered by silver followed by carbon (C) and oxygen (O). The occurrences of carbon and oxygen peaks may be due to the biological macromolecules like proteins and enzymes present on the surface of AgNPs.
Functional
FTIR analysis was performed in order to gain functional knowledge about the biological macromolecules (that possibly act as reducing/stabilizing agent) present in the culture supernatant of Stenotrophomonas sp. BHU-S7. The FTIR spectrum recorded from the dried powder of AgNPs is shown in Fig. 4b. The peaks near 3421 cm−1 and 2901 cm−1 were assigned to N-H stretching and C–H stretching, respectively. The peak at 1627 cm−1 corresponds to amide I, arising due to carbonyl stretch in enzymes and proteins. The broad peak at 1020 to 1250 cm−1 corresponds to aliphatic C–N stretching vibrations of the amines. The FTIR spectrum revealed that the carbonyl group of enzymes and proteins has the stronger ability to act as stabilizer by binding to the surface of the AgNPs. This result correlated well with the previous findings49,57. Altogether, FTIR spectroscopic analysis suggests that the release of extracellular proteins in the culture supernatant of Stenotrophomonas sp. BHU-S7 could possibly perform dual functions of synthesis and stabilization of AgNPs in the aqueous reaction medium.
Thermal
Thermal stability of the biosynthesized AgNPs was examined by TGA where temperature aided decomposition/desorption behavior of the particles is studied59. It has been observed from TGA plot that total dominant weight loss of 37.87% occurred in two steps in the temperature range of 0 °C to 700 °C while 62.13% of residual weight remained in the sample (Fig. 4c). The first step weight loss corresponding to 9.91% was observed at ~100 to 200 °C and the second step weight loss (27.96%) appeared in the temperature range of ~300–450 °C. The first step weight loss can be attributed to evaporation of water molecules adsorbed on the surface of AgNPs whereas second step weight loss is due to desorption of bioorganic components such as proteins and enzymes present in AgNPs sample.
Proficiency of biosynthesized AgNPs by Stenotrophomonas sp. BHU-S7 as nanofungicide against phytopathogens
AgNPs being the most effective and popular antimicrobial agent have capacity to kill nearly 650 types of pathogenic microbes including bacteria, fungi, viruses etc. within 30 minute60. The broad spectrum antimicrobial potency of AgNPs is a well-documented fact and has been widely described in many reports61,62,63,64,65. Similarly, several lines of evidence also describe potential antagonistic activity of AgNPs against diverse range of phytopathogenic fungi66,67,68,69. The idea of using biosynthesized AgNPs using biological agents for efficient management of plant diseases has gained noteworthy attention due to the fact that this biosynthesis process involves non-toxic and low cost biomolecules26. Moreover, biosynthesized AgNPs being equally competent to those nanoparticles generated by physical and chemicals methods provide safe and environment friendly approach for plant disease management. In context to this, several reports have successfully assessed the strong antifungal activity of biosynthesized AgNPs against phytopathogens24,25,26,31,32,70.
As pointed above, the present investigation emphasizes the sturdy impact of AgNPs biosynthesized using agricultural isolate of Stenotrophomonas sp. on some devastating plant pathogens such as A. alternata, C. lunata, B. sorokiniana and S. rolfsii. We examined the probable impact of biosynthesized AgNPs on conidial germination of foliar phytopathogens A. alternata, C. lunata and B. sorokiniana as this is the main disease causing step. Our data lucidly depict significant complete inhibition in conidial germination at 2, 4 and 10 μg/ml concentration under in vitro condition. While, on the contrary 100% conidial germination was observed in control set. In AgNO3 control set 20–30% inhibition was reported indicating superiority of AgNPs over AgNO3 in controlling plant diseases (Fig. 5a,b,c). The successful conidial germination over leaf surfaces is the main disease determining step leading to development of serious foliar diseases71. Hence, AgNPs mediated inhibition in conidial germination of such phytopathogens at very low concentration is of great importance and can be successfully applied for controlling devastating foliar diseases such as leaf spot, sheath blight, spot blotch etc.24,70. Additionally, we also examined the biocontrol potential of biosynthesized AgNPs against S. rolfsii, a soil-borne plant pathogen causing serious diseases on a variety of horticultural and agricultural crops. The efficient management of this soil borne plant pathogen is challenging and difficult as well because of its long term survival in soil due to presence of sclerotia72. Moreover, these hard structured sclerotia are well adapted for survival under stressed environment and resistant to physical and chemical degradation due to melanin accumulation in the outer membrane73. Therefore, we were more interested in studying the inhibitory impact of biosynthesized AgNPs on sclerotia germination. Our result clearly indicated that AgNPs treatment for 24 h completely reduced the sclerotia germination in PDA medium (Fig. 5d) while there was partial inhibition of sclerotia germination after 12 h treatment with AgNPs (data not shown). Exciting results were also obtained in soil system where noticeable reduction in sclerotia germination occurred after AgNPs treatment while in control set sclerotia started germinating after 24 h of incubation (Fig. 5e).
AgNPs generate highly reactive Ag+ ions which is actually responsible for its antimicrobial activity. Indeed, processes such as penetration into microbial cell, disruption of transport system, accumulation of Ag+ ions, and production of reactive oxygen species are the definite modes of actions of AgNPs responsible for its cidal effect on microbes74,75. Hence, it can be hypothesized that inhibitory effect of biosynthesized AgNPs on these phytopathogens is mainly attributed to one of these mechanisms. However, for verifying the exact mechanism a detail study is required.
Biosynthesized AgNPs mediated suppression of collar rot of chickpea
S. rolfsii causes collar rot disease in chickpea leading towards 50–95% mortality at seedling stage. The disease is more prevalent in attacking chickpea seedlings (maximum upto 4–6 weeks after sowing) under favorable condition i.e. high soil moisture and warmer climate76. The distinctive disease symptom is characterized by rotting of collar region due to development of whitish mycelial coating followed by yellowing and further collapsing of young seedlings. In the present investigation, greenhouse experiment revealed that AgNPs treatment resulted into suppression of collar rot of chickpea. No characteristic disease symptoms developed at the site of infection i.e. collar region after 24 h of pathogen challenge. Interestingly, chickpea plants looked as green and healthy as in control (without pathogen challenge) set. While in contrary, 100% mortality was observed in pathogen challenged set where plants got collapsed as a result of rotting at collar region after 48 h of pathogen inoculation (Fig. 6). These findings clearly indicated that AgNPs treatment provided protection to chickpea plants against Sclerotium rolfsii by hindering sclerotia germination. Our results represent direct evidence of interaction between sclerotia and biosynthesized AgNPs suggesting that reduced sclerotia germination is actually responsible for diminished inoculum potential and disease evidence. Furthermore, it can be hypothesized that such effects of AgNPs might be associated with disruption of sclerotial rind due to Ag+ penetration followed by their toxic accumulation inside the cell.
Defense response induced by phenolics in AgNPs treated chickpea against S. rolfsii infection
Phenolic compounds are known to confer disease resistance in plants against pathogen attack. Several mechanisms are involved in phenolic compounds mediated plant defense response that primarily includes building physical/chemical barrier through production of phytoanticipins (preformed) and phytoalexins (induced) and triggering signaling molecules for defense gene induction either locally or systemically77,78,79,80,81. It is well documented that diverse array of phenolic acids and flavonoids (phenolic acids derivatives) are synthesized in plants through phenylpropanoid pathway, by products of monolignol pathway and breakdown products of lignin78,82. Redeeming the immense importance of phenolics in plant-microbe interactions, we attempted to corroborate qualitative and quantitative differences in phenolics in response to S. rolfsii infection and biosynthesized AgNPs treatment.
The phenolic profiles revealed quantitative variations in phenolic compounds among the three treatments (Fig. 7a). The result indicated that most of the phenolic compounds were maximally induced in S. rolfsii challenged chickpea plants followed by maximal induction of ferulic acid and myricetin in AgNPs treated challenged (SR + AgNPs) plants. In general, the pattern of phenolic acids induction was observed to be as SR control >SR + AgNPs> Control (Table 1). The maximally induced phenolics in SR challenged plants were found to be shikimic acid (1170 μg/g), gallic acid (53.3 μg/g), syringic acid (118.4 μg/g), p-coumaric acid (232 μg/g), salicylic acid (547 μg/g), abscisic acid (11.7 μg/g), querecetin (46.2 μg/g), kaempferol (324 μg/g). Interestingly, many fold increase in kaempferol (324 μg/g) was reported in pathogen challenged plants as compared to control (10.05 μg/g) and SR + AgNPs (53.5 μg/g) plants. Apart from this, pathogen challenged chickpea plants treated with AgNPs experienced induction of ferulic acid (206 μg/g) and myricetin (121 μg/g) as compared to control and SR control plants. This marked differences in phenolics content among the treatments demonstrated that upon exposure to S. rolfsii, the chickpea plants felt stress and exhibited maximum induction of phenolic compounds resulted due to plant’s inherent immunity response in order to fight against the biotic stress. The maximally induced phenolic acids are known to exhibit antioxidant and antimicrobial property83,84. Alternatively, the direct contact between AgNPs and sclerotia weakened further progression and invasion of pathogen inside the host plant in SR + AgNPs treatment. Consequently, plants sensed relatively lesser stress and hence we observed not much difference in phenolic compounds between control and SR + AgNPs treatment. However, when compared with SR challenged plants, the increased induction of ferulic acid and myricetin was observed in SR + AgNPs treatment. Antioxidant property of ferulic acid and myricetin has been well documented in earlier reports85,86,87,88. Furthermore, ferulic acid is known to provide cell wall rigidity by building crosslink between polysaccharides and lignin89,90,91. These reports strongly indicate the potential role played by ferulic acid and myricetin against pathogen attack by making physical barrier and scavenging effect in response to free radicals generated upon exposure to S. rolfsii.
AgNPs mediated impact on lignification and H2O2 production in chickpea and their role in plant defense
Lignin is an important cell wall component in vascular plants that structurally strengthen and provide rigidity to the cell wall via cross linking with polysaccharides92,93. Lignification of plant tissue has been evidenced as an important mechanism of plant defense against pathogen attack by building a physical barrier to inhibit pathogen invasion inside the host plant cell94. Therefore, in the present investigation we attempted to examine the impact of AgNPs treatment on lignification pattern following pathogen attack. Interestingly, maximum lignification was observed in S. rolfsii challenged (SR control) chickpea plants followed by moderate lignification in SR + AgNPs treatment and least in control unchallenged plants. This differential response among the treatments resulted due to activation of plant’s inherent immunity response towards pathogen challenge and AgNPs treatment. Our findings point to positive correlation between increased lignification and severe stress condition95. Enhanced lignin deposition in cell wall in order to reduce the growth or confine the invading pathogen has been well described in earlier reports96,97,98. Despite of this very well-known fact, the direct impact of AgNPs on plant lignification pattern is a largely unexplored aspect. In relation to this, our study explores the probable impact of AgNPs on plant defense response towards pathogen attack. We evidenced that direct interaction between AgNPs and S. rolfsii gave rise to relatively reduced lignin deposition attributed to diminished pathogenicity as compared to SR control. Undoubtedly, it appears that AgNPs potentially exerted its toxic effect on S. rolfsii and hence SR + AgNPs treated plants experienced relatively lesser stress and displayed modification in defense response. The almost similar trend of lignification pattern in control and SR + AgNPs treatment revealed immense role of AgNPs in providing protection to chickpea plants against pathogen attack (Fig. 7b).
Apart from this, the strong evidence for AgNPs mediated protection to chickpea plants against S. rolfsii infection is well determined by studying in situ accumulation of H2O2 in plants using DAB staining method. In the present study, maximum number of brown spots indicative of H2O2 production appeared in pathogen challenged (SR control) chickpea leaves. Remarkably, the area and intensity of brown spots were highest due to S. rolfsii infection. In contrast to this, unchallenged control plants and SR + AgNPs treated plants displayed similar trend of brown coloration in leaves. It is important to note that the intensity of brown staining was relatively very low in these two treatments as compared to SR control (Fig. 7c). It is evident from earlier reports that H2O2 being the most stable reactive oxygen species (ROS) is thought to regulate plant development as well as stress response99,100. Moreover, H2O2 production in plants is indicative of an early response towards pathogen attack101. The results presented here strongly suggest that H2O2 production increased tremendously following S. rolfsii attack in SR control treatment. This elevated level is attributed to early response of chickpea plants to stress condition imposed by S. rolfsii by triggering its innate defense response. Plants accumulate higher level of ROS under stressed environment however, at low level they act as signal for triggering other defense response whereas their excessive accumulation lead to oxidative damage to protein, lipid and DNA resulting into plant death102,103. In relation to this, it can be hypothesized that collapsing of chickpea plants following S. rolfsii infection could be due to adverse impact of excessive H2O2 production on plant growth and other metabolic activity including lipid peroxidation, oxidative damage etc. On other hand, comparatively reduced level of H2O2 production in SR + AgNPs treated plants provides convincing evidence of important role played by AgNPs in providing protection to chickpea plants against S. rolfsii. It indicated that AgNPs treated plants are well equipped to counteract with pathogen challenge by increasing the activity of antioxidant enzymes that reduced the level of ROS and further improved stress tolerance.
Conclusion
The data presented in this article opens new perspective for efficient agricultural implications of bacterial strain Stenotrophomonas sp. that has not been attempted before. This study concerns the broad range antifungal properties of AgNPs biosynthesized using agricultural isolate of Stenotrophomonas sp. BHU S-7 (MTCC 5978), against devastating foliar and soil-borne phytopathogens. The biosynthesized AgNPs have been shown to adversely affect pathogenic propagules such as conidia and sclerotia leading to their reduced germination that ultimately diminished the future possibility of disease incidence. Additionally, the biosynthesized AgNPs exhibited successful management of chickpea collar rot disease caused by S. rolfsii under greenhouse condition. It is important to note that apart from in vitro studies, we made attempt to decipher the proficiency of biosynthesized AgNPs in natural environment for successful practical implication of this work in near future. The biosynthesized AgNPs mediated efficient management of collar rot disease was believed to be operated by many mechanisms. The reduced sclerotia germination, induction of phenolic acids, altered lignification and H2O2 production was the most reasonable explanation of the protective impact of AgNPs on chickpea against S. rolfsii infection (Fig. 8). The almost similar trend of phenolic profiling, stem lignin deposition and H2O2 production in unchallenged control and SR + AgNPs treatments revealed that AgNPs treated plants experienced lesser stress and are better equipped to cope with pathogen challenge. It also explains that AgNPs treatment helped in reducing the oxidative stress by enhancing the activity of antioxidants enzymes as evident by lowered level of H2O2 production in comparison to its excessive content in pathogen challenged plants. However, further investigations on studying the level of different antioxidant enzymes are required to support the proposed detail of mechanism. Altogether, the evidence achieved in the present study strengthen the perspective of robust and conceivable applications of AgNPs biosynthesized using greener approach and environment friendly Stenotrophomonas sp. BHU-S7 in agricultural sector. Moreover, we have gained new insight about the future possibilities of agricultural isolate of Stenotrophomonas sp. particularly for environmental benefits. Rethinking the debate addressing distinction between beneficial and harmful Stenotrophomonas strains, we strongly emphasize the selection of agricultural isolates of Stenotrophomonas sp. which enable their successful applications in agriculture. This may also be explained by the fact that pathogenic and non-pathogenic Stenotrophomonas strains share completely different niches and are likely to be adapted to that environment.
Additional Information
How to cite this article: Mishra, S. et al. Potential of biosynthesized silver nanoparticles using Stenotrophomonas sp. BHU-S7 (MTCC 5978) for management of soil-borne and foliar phytopathogens. Sci. Rep. 7, 45154; doi: 10.1038/srep45154 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Hugh, R. & Ryschenko, E. Pseudomonas maltophilia, an Alcaligenes like species. J Gen Microbiol 26, 123–132 (1961).
Moore, E. et al. 16S rRNA gene sequence analyses and inter- and intrageneric relationship of Xanthomonas species and Stenotrophomonas maltophilia . FEMS Microbiol Lett 151, 145–153 (1997).
Palleroni, N. J. & Bradbury, J. F. Stenotrophomonas, a new bacterial genus for Xanthomonas maltophilia (Hugh 1980) Swings et al. 1983. Int J Syst Bacteriol 43, 606–609 (1993).
Hayward, A. C., Fegan, N., Fegan, M. & Stirling, G. R. Stenotrophomonas and Lysobacter: ubiquitous plant‐associated gamma‐proteobacteria of developing significance in applied microbiology. J Appl Microbiol 108(3), 756–770 (2010).
Berg, G., Roskot, N. & Smalla, K. Genotypic and phenotypic relationships between clinical and environmental isolates of Stenotrophomonas maltophilia . J Clin Microbiol 37, 3594–3600 (1999).
Ryan, R. P. et al. The versatility and adaptation of bacteria from the genus Stenotrophomonas . Nat Rev Microbiol 7(7), 514–525 (2009).
Binks, P. R., Nicklin, S. & Bruce, N. C. Degradation of RDX by Stenotrophomonas maltophilia PB1. Appl Environ Microbiol 61, 1813–1822 (1995).
Boonchan, S., Britz, M. L. & Stanley, G. A. Surfactant enhanced biodegradation of high molecular weight polycyclic aromatic hydrocarbons by Stenotrophomonas maltophilia . Biotechnol Bioeng 59, 482–494 (1998).
Ikemoto, S., Suzuki, K., Kaneko, T. & Komagata, K. Characterization of strains of Pseudomonas maltophilia which do not require methionine. Int J Syst Bacteriol 30, 437–447 (1980).
Suckstorff, I. & Berg, G. Evidence for dose-dependent effects on plant growth by Stenotrophomonas strains from different origin. J Appl Microbiol 95, 656–663 (2003).
Schmidt, C. S., Alavi, M., Cardinale, M., Müller, H. & Berg, G. Stenotrophomonas rhizophila DSM14405T promotes plant growth probably by altering fungal communities in the rhizosphere. Biol Fertil Soils 48, 947–960 (2012).
Berg, G., Knaape, C., Ballin, G. & Seidel, D. Biological control of Verticillium dahlia KLEB by naturally occurring rhizosphere bacteria. Arch Phytopathol Dis Prot 29, 249–262 (1994).
Kobayashi, D. Y., Gugliemoni, M. & Clarke, B. B. Isolation of chitinolytic bacteria Xanthomonas maltophilia and Serratia marcescens as biological control agents for summer patch disease of turf grass. Soil Biol Biochem 27, 1479–1487 (1995).
Nakayama, T., Homma, Y., Hashidoko, Y., Mitzutani, J. & Tahara, S. Possible role of xanthobaccins produced by Stenotrophomonas sp. strain SB-K88 in suppression of sugar beet damping-off disease. Appl Environ Microbiol 65, 4334–4339 (1999).
Dunne, C., Moenne-Loccoz, Y., de Bruijn, F. J. & O’Gara, F. Overproduction of an inducible extracellular serine protease improves biological control of Pythium ultimum by Stenotrophomonas maltophilia strain W81. Microbiology 146, 2069–2078 (2000).
Zhang, Z., Yuen, G. Y., Sarath, G. & Penheiter, A. R. Chitinases from the plant disease biocontrol agent, Stenotrophomonas maltophilia C3. Phytopathology 91, 204–211 (2001).
Kai, M., Effmert, U., Berg, G. & Piechulla, B. Volatiles of bacterial antagonists inhibit mycelial growth of the plant pathogen Rhizoctonia solani . Arch Microbiol 187, 351–360 (2007).
Wolf, A., Fritze, A., Hagemann, M. & Berg, G. Stenotrophomonas rhizophila sp. nov., a novel plant-associated bacterium with antifungal properties. Int J Syst Evol Microbiol 52, 1937–1944 (2002).
Berg, G. & Martinez, J. L. Friends or foes: can we make a distinction between beneficial and harmful strains of the Stenotrophomonas maltophilia complex? Front Microbiol 6, 241 (2015).
Berg, G. et al. Biocontrol and osmoprotection for plants under salinated conditions. In Molecular Microbial Ecology of the Rhizosphere: Volume 1 & 2 (Ed. de Bruijn, F. J. ), John Wiley & Sons, Inc., Hoboken, NJ, USA, 561–573 (2013).
Hagemann, M. et al. The plant-associated bacterium Stenotrophomonas rhizophila expresses a new enzyme for the synthesis of the compatible solute glucosylglycerol. J Bacteriol 190(17), 5898–5906 (2008).
Egamberdieva, D. et al. Bacteria able to control foot and root rot and to promote growth of cucumber in salinated soils. Biol Fertility Soils 47(2), 197–205 (2011).
Lugtenberg, B. & Kamilova, F. Plant-growth-promoting rhizobacteria. Annu Rev Microbiol 63, 541–556 (2009).
Mishra, S. et al. Biofabricated silver nanoparticles act as a strong fungicide against Bipolaris sorokiniana causing spot blotch disease in wheat. PLOS One 9(5), e97881 (2014a).
Mishra, S., Singh, A., Keswani, C. & Singh, H. B. Nanotechnology: Exploring potential application in agriculture and its opportunities and Constraints. Biotech Today 4(1), 9–14 (2014b).
Mishra, S. & Singh, H. B. Biosynthesized silver nanoparticles as a nanoweapon against phytopathogens: exploring their scope and potential in agriculture. Appl Microbiol Biotechnol 99, 1097–1107 (2015a).
Mishra, S. et al. Microbial nanoformulation: Exploring potential for coherent nanofarming. In The Handbook of Microbial Resources (Eds Vijai K. Gupta et al.), CABI, pp, 107–120 (2016).
Li, X., Xu, H., Chen, Z. S. & Chen, G. Biosynthesis of nanoparticles by microorganisms and their applications. J Nanomater 2011, 8 (2011).
Carma, R. S. Greener approach to nanomaterials and their sustainable applications. Curr Opin Chem Eng 1, 123–128 (2012).
Hulkoti, N. I. & Taranath, T. C. Biosynthesis of nanoparticles using microbes-A review. Colloids and Surf B Biointerfaces 121, 474–483 (2014).
Krishnaraj, C., Ramachandran, R., Mohan, K. & Kalaichelvan, P. T. Optimization for rapid synthesis of silver nanoparticles and its effect on phytopathogenic fungi. Spectrochim Acta A Mol Biomol Spectrosc 93, 95–99 (2012).
Gopinath, V. & Velusamy, P. Extracellular biosynthesis of silver nanoparticles using Bacillus sp. GP-23 and evaluation of their antifungal activity towards Fusarium oxysporum . Spectrochim Acta A Mol Biomol Spectrosc 106, 170–174 (2013).
Lee, K. J. et al. Synthesis of silver nanoparticles using cow milk and their antifungal activity against phytopathogens. Mater Lett 105, 128–131 (2013).
Paulkumar, K. et al. Piper nigrum leaf and stem assisted green synthesis of silver nanoparticles and evaluation of its antibacterial activity against agricultural plant pathogens. The Scientific World Journal 2014, Article ID 829894, 9 pages (2014).
Raliya, R., Nair, R., Chavalmane, S., Wang, W. N. & Biswas, P. Mechanistic evaluation of translocation and physiological impact of titanium dioxide and zinc oxide nanoparticles on the tomato (Solanum lycopersicum L.) plant. Metallomics 7(12), 1584–1594 (2015).
Mishra, S. & Singh, H. B. Preparation of biomediated metal nanoparticles. Indian Patent 201611003248, Patent filed (2016).
Nangia, Y., Wangoo, N., Goyal, N., Shekhawat, G. & Suri, C. R. A novel bacterial isolate Stenotrophomonas maltophilia as living factory for synthesis of gold nanoparticles. Microb Cell Fact 8(1), 1 (2009).
Malhotra, A. et al. Biosynthesis of gold and silver nanoparticles using a novel marine strain of Stenotrophomonas . Bioresour Technol 142, 727–731 (2013).
Oves, M. et al. Antibacterial and cytotoxic efficacy of extracellular silver nanoparticles biofabricated from chromium reducing novel OS4 strain of Stenotrophomonas maltophilia . PloS One 8(3), e59140 (2013).
Mishra, S. & Nautiyal, C. S. Reducing the allelopathic effect of Parthenium hysterophorus L. on wheat (Triticum aestivum) by Pseudomonas putida . Plant Growth Regul 66, 155–165 (2012).
Singh, B. R. et al. Mycofabricated biosilver nanoparticles interrupt Pseudomonas aeruginosa quorum sensing systems. Sci Rep 5 (2015).
Ashraf, J. M., Ansari, M. A., Khan, H. M., Alzohairy, M. A. & Choi, I. Green synthesis of silver nanoparticles and characterization of their inhibitory effects on AGEs formation using biophysical techniques. Sci Rep 6 (2016).
Niranjan, A. et al. Identification and quantification of heterologous compounds parthenin and organic acids in Parthenium hysterophorus L. using HPLC-PDA-MS-MS. Anal Lett 46(1), 48–59 (2013).
Thordal-Christensen, H., Zhang, Z., Wei, Y. & Collinge, D. B. Subcellular localization of H2O2 in plants, H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J 11, 1187–1194 (1997).
Jensen, W. A. Botanical histochemistry: principles and practices London, WH Freeman and Co. (1962).
Sadowski, Z., Maliszewska, I. H., Grochowalska, B., Polowczyk, I. & Ozlecki, T. Synthesis of silver nanoparticles using microorganisms. Mater Sci-Poland 26, 419–424 (2008).
Kalimuthu, K., Suresh Babu, R., Venkataraman, D., Bilal, M. & Gurunathan, S. Biosynthesis of silver nanocrystals by Bacillus licheniformis . Colloids Surf B Biointerfaces 65, 150–153 (2008).
Mulvaney, P. Surface plasmon spectroscopy of nanosized metal particles. Langmiur 12, 788–800 (1996).
Balaji, D. S. et al. Extracellular biosynthesis of functionalized silver nanoparticles by strains of Cladosporium cladosporioides fungus. Colloids Surf B Biointerfaces 68(1), 88–92 (2009).
Durán, N., Marcato, P. D., Alves, O. L., De Souza, G. & Esposito, E. Mechanistic aspects of biosynthesis of silver nanoparticles by several Fusarium oxysporum strains. J Nanobiotechnol 3(8), 1–7 (2005).
Sastry, M., Mayya, K. S. & Bandyopadhyay, K. pH Dependent changes in the optical properties of carboxylic acid derivatized silver colloidal particles. Colloids Surf A Physicochem Eng Asp 127, 221–228 (1997).
Jain, P. K., Huang, X., El-Sayed, I. H. & El-Sayed, M. A. Noble metals on the nanoscale: optical and photothermal properties and some applications in imaging, sensing, biology, and medicine. Acc Chem Res 41(12), 1578–1586 (2008).
Link, S. & El-Sayed, M. A. Spectral properties and relaxation dynamics of surface plasmon electronic oscillations in gold and silver nanodots and nanorods. J Phys Chem B 103(40), 8410–8426 (1999).
Dorofeev, G. A., Streletskii, A. N., Povstugar, I. V., Protasov, A. V. & Elsukov, E. P. Determination of nanoparticle sizes by X-ray diffraction. Colloid J 74(6), 675–685 (2012).
Chen, C., Yu, B., Liu, J., Dai, Q. & Zhu, Y. Investigation of ZnO films on Si <111> substrate grown by low energy O+ assisted pulse laser deposited technology. Mater Lett 61(14), 2961–2964 (2007).
Sathiya, C. K. & Akilandeswari, S. Fabrication and characterization of silver nanoparticles using Delonix elata leaf broth. Spectrochim Acta A Mol Biomol Spectrosc 128, 337–341 (2014).
Kasthuri, J., Veerapandian, S. & Rajendiran, N. Biological synthesis of silver and gold nanoparticles using apiin as reducing agent. Colloids Surf B Biointerfaces 68(1), 55–60 (2009).
Magudapathy, P. et al. Electrical transport studies of Ag nanocrystallites embedded in glass matrix. Physica B Condens Matter 299, 142 (2001).
Ankireddy, K. et al. Thermal analysis of silver nanoparticles for flexible printed antenna fabrication. J Appl Phys 114(12), 124303 (2013).
Shahrokh, S. & Emtiazi, G. Toxicity and unusual biological behavior of nanosilver on gram positive and negative bacteria assayed by microtiter-plate. Eur J Biol Sci 1, 28–31 (2009).
Kim, J. S. et al. Antimicrobial effects of silver nanoparticles. Nanomedicine Nanotechnol Biol Med 3, 95–101 (2007).
Ma, X., Geiser-Lee, J., Deng, Y. & Kolmakov, A. Interactions between engineered nanoparticles (ENPs) and plants: phytotoxicity, uptake and accumulation. Sci Total Environ 408, 3053–3061 (2010).
Lemire, J. A., Harrison, J. J. & Turner, R. J. Antimicrobial activity of metals: mechanisms, molecular targets and applications. Nat Rev Microbiol 11, 371–384 (2013).
Kora, A. J. & Sashidhar, R. B. Antibacterial activity of biogenic silver nanoparticles synthesized with gum ghatti and gum olibanum: a comparative study. J Antibiot 68, 88–97 (2015).
Wong, M. S. et al. Antibacterial property of Ag nanoparticle-impregnated N-doped titania films under visible light. Sci Rep 5, (2015).
Park, H. J., Kim, S. H., Kim, S. J. & Choi, S. H. A new composition of nanosized silica-silver for control of various plant diseases. Plant Pathol J 22, 295–302 (2006).
Kim, S. W. et al. An in vitro study of the antifungal effect of silver nanoparticles on oak wilt pathogen Raffaelea sp. J Microbiol Biotechnol 19, 760–764 (2009).
Lamsal, K. et al. Application of silver nanoparticles for the control of Colletotrichum species in vitro and pepper anthracnose disease in field. Mycobiology 39(3), 194–199 (2011).
Kim, S. W. et al. Antifungal effects of silver nanoparticles (AgNPs) against various plant pathogenic fungi. Mycobiology 40, 53–58 (2012).
Mishra, S. & Singh, H. B. Silver nanoparticles mediated altered gene expression of melanin biosynthesis genes in Bipolaris sorokiniana . Microbiol Res 172, 16–18 (2015b).
Bradley, D. J., Gilbert, G. S. & Parker, I. M. Susceptibility of clover species to fungal infection: the interaction of leaf surface traits and environment. Am J Bot 90(6), 857–864 (2003).
Boyle, L. W. The ecology of Sclerotium rolfsii with emphasis on the role of saprophytic media. Symposium on Sclerotium rolfsii. Phytopathology 51, 117–119 (1961).
Chet, I. Ultrastructural basis of sclerotial survival in soil. Microb Ecol 2(3), 194–200 (1975).
Morones, J. R. et al. The bactericidal effect of silver nanoparticles. Nanotechnology 16, 2346–2353 (2005).
Hwang, E. T. et al. Analysis of the toxic mode of action of silver nanoparticles using stress-specific bioluminescent bacteria. Small 4, 746–750 (2008).
Singh, U. P., Sarma, B. K. & Singh, D. P. Effect of plant growth-promoting rhizobacteria and culture filtrate of Sclerotium rolfsii on phenolic and salicylic acid contents in chickpea (Cicer arietinum). Curr Microbiol 46(2), 0131–0140 (2003).
Dixon, R. A. & Paiva, N. L. Stress induced phenylpropanoid metabolism. Plant Cell 7, 1085–1097 (1995).
Dixon, R. A. Natural products and plant disease resistance. Nature 411, 843–847 (2001).
Treutter, D. Significance of flavonoids in plant resistance and enhancement of their biosynthesis. Plant Biol 7, 581–591 (2005).
Mandal, S. M., Chakraborty, D. & Dey, S. Phenolic acids act as signaling molecules in plant-microbe symbioses. Plant Signal Behav 5(4), 359–368 (2010).
Mishra, S. et al. Harnessing plant-microbe interactions for enhanced protection against phytopathogens. In Arora, N. K. (Ed.) Plant Microbes Symbiosis: Applied Facets Springer, India, pp. 111–125 (2015c).
Croteau, R., Kutchan, T. M. & Lewis, N. G. Natural products (secondary metabolites). In Buchanan et al. (eds) Biochemistry and molecular biology of plants American Society of Plant Physiologists, Rockville MD 1250–318 (2000).
Shetty, R. et al. Silicon-induced changes in antifungal phenolic acids, flavonoids, and key phenylpropanoid pathway genes during the interaction between miniature roses and the biotrophic pathogen Podosphaera pannosa . Plant Physiol 157(4), 2194–2205 (2011).
Jain, A., Singh, A., Singh, S. & Singh, H. B. Phenols enhancement effect of microbial consortium in pea plants restrains Sclerotinia sclerotiorum . Biol Control 89, 23–32 (2015).
Kikuzaki, H., Hisamoto, M., Hirose, K., Akiyama, K. & Taniguchi, H. Antioxidant properties of ferulic acid and its related compounds. J Agric Food Chem 50(7), 2161–2168 (2002).
Ou, S. & Kwok, K. C. Ferulic acid: pharmaceutical functions, preparation and applications in foods. J Sci Food Agric 84, 1261–1269 (2004).
Shimoji, H. & Yamasaki, H. Inhibitory effects of flavonoids on alternative respiration of plant mitochondria. Biol Plant 49(1), 117–119 (2005).
Rosa, N. N., Dufour, C., Lullien-Pellerin, V. & Micard, V. Exposure or release of ferulic acid from wheat aleurone: impact on its antioxidant capacity. Food Chem 141(3), 2355–2362 (2013).
Strack, D. Metabolism of hydroxycinnamic acid conjugates. Bull Liaison Groupe Polyphen 15, 55–64 (1990).
Rosazza, J. P. N., Huang, Z., Dostal, L., Volm, T. & Rousseau, B. Review: biocatalytic transformations of ferulic acid: an abundant aromatic natural product. J Ind Microbiol 15(6), 457–471 (1995).
Kumar, N. & Pruthi, V. Potential applications of ferulic acid from natural sources. Biotechnol Rep 4, 86–93 (2014).
Sattler, S. E. & Funnell-Harris, D. L. Modifying lignin to improve bioenergy feed stocks: strengthening the barrier against pathogens? Front Plant Sci 4, 70 (2013).
Malinovsky, F. G., Fangel, J. U. & Willats, W. G. The role of the cell wall in plant immunity. Plant Cell Wall in Pathogenesis, Parasitism and Symbiosis 38 (2015).
Vance, C. P., Kirk, T. K. & Sherwood, R. T. Lignification as a mechanism of disease resistance. Annu Rev Phytopathol 18, 259–288 (1980).
Lee, B. R. et al. Peroxidases and lignification in relation to the intensity of water-deficit stress in white clover (Trifolium repens L.). J Exp Bot 58, 1271–1279 (2007).
Hudgins, J. W., Christiansen, E. & Franceschi, V. R. Induction of anatomically based defense responses in stems of diverse conifers by methyl jasmonate: a phylogenetic perspective. Tree Physiol 24, 251–264 (2004).
Menden, B., Kohlhoff, M. & Moerschbacher, B. M. Wheat cells accumulate a syringyl-rich lignin during the hypersensitive resistance response. Phytochemistry 68, 513–520 (2007).
Kissoudis, C., van de Wiel, C., Visser, R. G. & van der Linden, G. Enhancing crop resilience to combined abiotic and biotic stress through the dissection of physiological and molecular crosstalk. Front Plant Sci 5, 207 (2014).
Bienert, G. P. et al. Specific aquaporins facilitate the diffusion of hydrogen peroxide across membranes. J Biol Chem 282(2), 1183–1192 (2007).
Van Breusegem, F., Bailey-Serres, J. & Mittler, R. Unraveling the tapestry of networks involving reactive oxygen species in plants. Plant Physiol 147, 978–984 (2008).
Vanacker, H., Carver, T. L. & Foyer, C. H. Early H2O2 accumulation in mesophyll cells leads to induction of glutathione during the hyper-sensitive response in the barley-powdery mildew interaction. Plant Physiol 123(4), 1289–1300 (2000).
Apel, K. & Hirt, H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55, 373–399 (2004).
Liu, Y. H., Offler, C. E. & Ruan, Y. L. A simple, rapid, and reliable protocol to localize hydrogen peroxide in large plant organs by DAB-mediated tissue printing. Front Plant Sci 5 (2014).
Acknowledgements
Sandhya Mishra is grateful to the Department of Science and Technology, Government of India, New Delhi, for financial assistance under SERB-Start Up Research Grant (Young Scientist) Scheme (YSS/2015/000082).
Author information
Authors and Affiliations
Contributions
H.B.S. conceived and designed the experiments. S.M. and B.R.S. carried out the experiments and analyzed the data. H.B.S., S.M., B.R.S. and A.H.N. wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Mishra, S., Singh, B., Naqvi, A. et al. Potential of biosynthesized silver nanoparticles using Stenotrophomonas sp. BHU-S7 (MTCC 5978) for management of soil-borne and foliar phytopathogens. Sci Rep 7, 45154 (2017). https://doi.org/10.1038/srep45154
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep45154
- Springer Nature Limited
This article is cited by
-
Silver nanoparticles as potential fungicide against rice brown spot: physiological and biochemical responses in plants
Tropical Plant Pathology (2024)
-
Bio-Synthesized Silver Nanoparticles Using Cannabis Sativa Seed Extracts and Its Anticancer Effects
Plasmonics (2024)
-
Tiny but mighty: metal nanoparticles as effective antimicrobial agents for plant pathogen control
World Journal of Microbiology and Biotechnology (2024)
-
A Minor Groove Binder with Significant Cytotoxicity on Human Lung Cancer Cells: The Potential of Hesperetin Functionalised Silver Nanoparticles
Journal of Fluorescence (2024)
-
Nanomicrobiology: Emerging Trends in Microbial Synthesis of Nanomaterials and Their Applications
Journal of Cluster Science (2023)