Abstract
Human cancers frequently display substantial intra-tumoural heterogeneity in virtually all distinguishable phenotypic features, such as cellular morphology, gene expression, and metastatic potential. In order to investigate tumour heterogeneity in myxofibrosarcoma, we established a novel myxofibrosarcoma cell line with two well defined sub-clones named MUG-Myx2a and MUG-Myx2b. The parental tumour tissue and both MUG-Myx2 cell lines showed the same STR profile. The fact that MUG-Myx2a showed higher proliferation activity, faster migration and enhanced tumourigenicity was of particular interest. NGS mutation analysis revealed corresponding mutations in the FGFR3, KIT, KDR and TP53 genes. In contrast, the MUG-Myx2a cell lines showed an additional PTEN mutation. Analysis of CNV uncovered a highly aberrant karyotype with frequent losses and gains in the tumour sample. The two MUG-Myx2 cell lines share several CNV features of the tumour tissue, while some CNVs are present only in the two cell lines. Furthermore, certain CNV gains and losses that are exclusive to either MUG-Myx2a or MUG-Myx2b, distinguish the two cell lines. As it is currently not possible to purchase two different sarcoma cell lines derived from the same patient, the novel myxofibrosarcoma cell lines MUG-Myx2a and MUG-Myx2b will be useful tools to study pathogenesis, tumour heterogeneity and treatment options.
Similar content being viewed by others
Introduction
Myxofibrosarcoma (MFS) is the most common sarcoma in elderly patients and is characterised histologically by a multinodular growth pattern and variably prominent myxoid stroma. The tumour is mainly composed of spindle cells with variable cytologic atypia accentuated along curvilinear vessels1. Clinically, increasing grades and stages of the tumours are frequently seen in MFS after local recurrences, which may eventually lead to metastatic diseases. Recurrence has been shown to occur despite repeated surgery involving wide-ranging local excisions and negative surgical margins1,2,3. MFS is characterized by non-specific cytogenetic aberrations that increase with grade, suggesting a multistep tumour progression model due to acquired genetic instability4. The mechanisms responsible for the coexistence of distinct sub-clones and the biological consequences of this coexistence remain poorly understood. MALDI imaging mass spectrometry demonstrated a high grade of intra-tumour heterogeneity in MFS5. Despite their histological resemblance, imaging mass spectrometry demonstrated that intra-tumour heterogeneity was a consistent feature in each of the MFS tumours studied, and revealed that different nodules exhibited high grade-/low grade-like biomolecular signatures. Hence, cytogenetic heterogeneity and clonal evolution seem to be common in myxoid sarcoma5,6. In addition, large-scale sequencing analyses of other solid cancers have identified extensive heterogeneity both between individual tumors and genetic intra-tumour heterogeneity7,8,9. Genomic instability generates a high level of intercellular genetic heterogeneity and has been linked to both drug resistance and poor prognosis in cancer, because therapeutic procedures rely on single tumour biopsy samples10,11,12.
In this context the availability of appropriate in vitro cell systems is of particular importance. Permanent cell lines derived from primary sarcomas offer the opportunity to study functional alterations in sarcoma biology. However, the supply of commercial sarcoma cell lines is extremely inadequat. In order to investigate this intra-tumour heterogeneity of MFS in vitro, we established two sub-clones of a primary MFS, named MUG-Myx2a and MUG-Myx2b. The new cell lines provide a model for tumour heterogeneity and allow comprehensive genetic and epigenetic characterization. Furthermore, because of their tumourigenicity, the cell lines are also suitable for in vivo experiments. The present study describes the establishment, various growth kinetics, migration activity, tumourigenicity, cytogenetic features and mutation analysis of the newly established MFS cell lines.
Results
Establishment of the novel MFS sub-clones MUG-Myx2a and MUG-Myx2b
Haematoxylin and eosin (HE)-stained slides of a 94-year old female patient revealed a G3 stage MFS tumour. The tumour was composed of morphologically heterogenous areas. Partially, the tumour showed abundant myxoid stroma with classic curvilinear tumour vessels. In addition, accentuated atypical tumour cells with spindle morphology were located around vascular structures, consistent with well differentiated tumour areas. Furthermore, the tumour showed a high grade tumour component composed of fascicles of pleomorphic tumour cells and brisk mitotic activity. Immunohistochemical (IHC) staining of the patient’s tumour revealed only focal SMA positivity, whereas the tumour was negative for Desmin, Caldesmon, S100, CD34, EMA, Melan A, and Pan-CK. (data not shown). After crushing and enzymatically digesting the tumour tissue, starting from the primary culture (passage 0) two different sub-clones were successfully expanded. As early as the beginning of cultivation, the clones named MUG-Myx2a and MUG-Myx2b differed in terms of their morphology. MUG-Myx2a and MUG-Myx2b showed elongated spindle-shaped cells, with large nuclei and prominent nucleoli, and grown attached to a substrate. It is notable, that throughout the entire duration of cultivation MUG-Myx2a cells were markedly more elongated and more spindle-shaped than MUG-Myx2b (Fig. 1A, B). The mesenchymal origin of the cells was confirmed by high vimentin expression (Fig. 1B). To demonstrate the identical origin of the cell lines, a short tandem repeat (STR) Power Plex® 16 analysis of the frozen primary parental tumour tissue, as well as MUG-Myx2a (passage 45) and MUG-Myx2b (passage 48) was performed. The original tissue and cell lines showed the same STR profile for the markers TH01, D21S11, Penta E, D5S818, D13S317, D7S820, CSF1PO, Penta D, Amelogenin, D8S1179, TPOX, and FGA. All values are summarised in Table 1.
(A) Morphological features of MUG-Myx2a and MUG-Myx2b cells at passage 1, 43, and 90. (B) Strong expression of Vimentin of MUG-Myx2a and MUG-Myx2b confirmed the mesenchymal origin of the tumour cell lines; nuclei were counterstained with DAPI. (C) MTS proliferation analysis revealed highly significant variations at all time points. Dynamic proliferation curves measured with the xCELLigence system showed (D) a faster growth rate and (E) a higher migration potential for MUG-Myx2a cells than the MUG-Myx2b cells. X axis represents CI (cell index) and y axis the time in hours.
MUG-Myx2a cells show higher proliferation, migration, and tumourigenicity
The growth behaviour of the two clones was investigated with the MTS assay after 1 to 6 days. The statistical comparability of the two clones revealed highly significant variations at all time points (day 1: p = 0.00064; day 2: p = 1.87E-06; day 3: 2.82E-17; day 6: 6.23E-06; n = 12) (Fig. 1C). In order to evaluate the real-time growth pattern and migration potential, cells were detected in quadruplicate with the xCELLigence System. Using the RTCA 1.2.1 software, the population doubling time of the cells was calculated. MUG-Myx2a cells displayed a lower doubling time (20 h vs 22 h) and a higher migration potential than MUG-Myx2b cells (Fig. 1D,E). After treatment with 0–25 μM doxorubicin, 0–25 μM Verinostat, and 0–25 nM bortezomib for 48 h, cell viability was measured by the MTS assay. Chemotherapeutic drugs inhibited cell growth in a dose-dependent manner in both cell lines (Fig. 2A–C). Nevertheless MUG-Myx2a cells showed a significant higher sensitivity to all three therapeutic drugs.
MUG-Myx2a (p49) and MUG-Myx2b (p52) cells successfully formed tumours in 3 of 5 transplanted mice. Small nodules were palpable 4 weeks after inoculation. After 8 weeks the animal were dissected. Tumour formation in mice was significantly higher when MUG-Myx2a cells were injected, in comparison to MUG-Myx2b (Fig. 3A). The success rate of growing MUG-Myx2a/b cells in nu/nu Foxn1 mice was 60%. Successful engraftment was determined by pathological examination of formalin-fixed, paraffin-embedded (FFPE) material from the tumour samples. Hematoxylin and eosin (HE)-stained FFPE mice tumours showed variation concerning the morphology of MUG-Myx2a (Fig. 3B) and MUG-Myx2b (Fig. 3C). In particular, MUG-Myx2b showed the characteristic prominent myxoid stroma of MFS.
The high mitotic rate and high proliferative index were confirmed by IHC with the proliferation marker Ki-67. Using the ImageScope software, Ki-67 positive and negative cells were quantified after IHC staining. MUG-Myx2a tumours from all mice displayed an increased proliferation level, compared to MUG-Myx2b tumours (Ki-67 positivity: 0.937 ± 0.033 vs 0.604 ± 0.185; n = 3). A representative staining of one pair of tumours is shown and summarized in Fig. 3D (MUG-Myx2a) and 3E (MUG-Myx2b).
Mutation Analysis by Next-Generation Sequencing (NGS)
Mutation Analysis of 50 genes by NGS revealed that the primary patient’s tumour tissue and MUG-Myx2b showed identical mutations in the FGFR3, KIT, KDR, and TP53 genes. Conversely, the MUG-Myx2a cell line showed the same mutations as MUG-Myx2b and the primary patient tumour, but had an additional mutation in the PTEN gene (Table 2).
Chromosomal Copy Number Variations (CNVs)
Low coverage whole genome sequencing was used to determine chromosomal gains and losses in the primary tumour tissue as well as in both MUG-Myx2 cell lines. The tumour tissue showed widespread gains and losses for several chromosomes (Fig. 4, outer ring). The MUG-Myx2 cell lines both showed gains for chromosomes 3, 6, 8, 14, and 17 as well as losses of chromosomes 3, 6, 9, and 13. Additionally, both cell lines showed gains for chromosomes 1, 3, and 6 and losses of chromosomes 8 and X. These alterations were not evident in the primary tumour tissue. MUG-Myx2a (Fig. 4, middle ring) showed losses of chromosome 4 and gains of chromosome 12, but these characteristics were not shared with the primary tumour, or the MUG-Myx2b cell line. On the other hand, MUG-Myx2b (Fig. 4, innermost ring) exhibited losses of chromosome 5, which were exclusive to this cell line. In addition to these substantial CNVs, we also determined the copy number state of 50 cancer genes, which were also tested for the presence of mutations. Both MUG-Myx2 cell lines showed a similar pattern of CNVs in these genes and shared the distinct loss of CDKN2a and MLH1, as well as a gain of FGFR1, with the primary tumor tissue (Fig. 5; Table 3). However, gain of the JAK3 and NRAS locus was only seen in the MUG-Myx2 cell lines.
Discussion
Myxofibrosarcoma is a malignant neoplasm with variably prominent myxoid stroma, cellular pleomorphism, and a distinct curvilinear vascular pattern. It represents the most common sarcoma in elderly patients, with a slight predominance in males1. In order to enable the investigation of tumour heterogeneity on cellular and molecular level, it is highly desirable to establish new human primary cell lines.
Clonal heterogeneity may be evident within single samples, but can also be observed between different tumour regions within the same primary site, or even between primary and metastatic sites, so-called “regional heterogeneity”13,14.
MUG-Myx2a and MUG-Myx2b cell lines showed the same STR profile and could be maintained in long-term cultures with a 20 or 22 h doubling time, respectively. Interestingly, the MUG-Myx2a showed a higher proliferation and migration ability than MUG-Myx2b. Additionally, compared to MUG-Myx2b, MUG-Myx2a cells showed a higher sensitivity to commonly used chemotherapeutic drugs.
Both cell lines successfully formed tumours in nu/nu Foxn1 mice, although the MUG-Myx2a cell line again showed higher proliferation capacity and thereby confirmed the growth analysis data. A nu/nu Foxn1 mouse model demonstrated, that both cell lines showed equally aggressive in vivo growth behaviour. Using an alternative mouse model, such asNSG mice, might show increased tumourigenicity.
A distinctive feature of cancer cells compared to normal cells is chromosome instability, which is proposed to be critical for the initiation of tumourigenesis15. For this reason, we investigated the genomic integrity of the MUG-Myx2 cell lines by low density whole genome sequencing. Gains in gene copy number can drive the expression of oncogenes, whereas decreased gene dosage by hemizygous and/or homozygous deletion may inactivate tumour suppressor genes16. The MUG-Myx2 cell lines match some, but not all chromosomal aberrations of the primary tumour. Interestingly, many features seem to be more pronounced in the cell lines, consistant with the notion, that the primary tumour contains several sub-clonal populations of cells which cumulatively contribute to the CNV phenotype of the primary tumour. However, the two cell lines might represent single sub-clones which share many features with the primary tumour, but have also accrued additional changes. In the CNV analysis of individual genes, the copy number state of the CDKN2a, MLH1, and FGFR1 genes is identical in the primary tumour and the cell lines. CDKN2a, also known as p16, has been described as frequently deleted in osteosarcomas17 whereas we found changes which are present in both the primary tumour and MUG-Myx2a/b. MLH1 is part of the DNA mismatch repair system and its loss could contribute to the widespread genetic changes seen in this tumour. FGFR1 amplification is a common feature of several tumour types and activates the cell cycle via the RAS pathway.
The recent development of NGS as a diagnostic tool in the clinical setting has enabled rapid, targeted sequencing of tumours to determine causative mutations. When combined with various selective capture approaches, NGS has allowed for the efficient simultaneous genetic analysis of a large number of candidate genes. Here, we applied polymerase chain reaction (PCR) based NGS to identify alterations to oncogenes and tumour suppressor genes. Our NGS data revealed, that both MUG-Myx2a and MUG-Myx2b cells possessed the p53 mutation R213Q, whereas, only MUG-Myx2a showed the PTEN R173H mutation.
p53 and PTEN are the two most frequently mutated tumour suppressors in human cancer. p53 plays a major role in regulating the response of mammalian cells to stress and damage, partially through transcriptional activation of genes involved in cell cycle control, DNA repair, senescence, angiogenesis, and apoptosis18. Furthermore, mutations in p53 have also been identified as the most common genetic alterations in sarcoma19. PTEN is involved in basic cellular functions including adhesion, migration, proliferation and cell survival20. Somatic mutations in PTEN are now known to cause tumourigenesis in a number of human tissues21. Both p53 and PTEN are involved in sustaining cellular homeostasis and complex regulatory interactions22,23. The combined inactivation pf p53 and PTEN could greatly accelerate tumour development24,25,26. Mutational analysis of p53 and PTEN in STS tissue samples revealed an incidence of 25.6% (22 out of 86) p53 mutations but only 2.3% (2 out of 86) PTEN mutations27. The PTEN R173H mutation is located in one of the three hotspot sites of the PTEN gene and has been reported in patients with PTEN associated tumors28. This mutation reduces PTEN activity by 50%29. The absence of this mutation from the primary tumor as well as the MUG-Myx2b cell line can either be explained by tumor heterogeneity, with MUG-Myx2a cells originating from a tumor sub-clone not present in the paraffin block sample used for analysis of the primary tumor, or by a secondary mutation during cell culture.
KDR (kinase insert domain receptor) is the human gene encoding vascular endothelial growth factor receptor 2 (VEGFR-2). It functions as the main mediator of VEGF-induced endothelial proliferation, survival, migration, tubular morphogenesis, and sprouting. Several studies have reported the genetic polymorphism of the KDR gene and the implicated risk of coronary artery diseases30,31. However, the clear role of individual KDR SNPs and their physiological functions in cancer progression and prognosis remains unknown. Mutations in the KIT receptor tyrosine kinase, which are commonly present in gastrointestinal stromal tumours (GISTs; 70–80% of all cases) are clustered in four exons32. The FGFR3 F386L polymorphism has been reported in association with low-grade tumours and early disease stage in prostate cancer33.
In conclusion, the well-characterized novel MFS cell lines MUG-Myx2a and MUG-Myx2b will be a useful tool to gain further insights into the pathogenesis and tumour heterogeneity of MFS and explore new therapy options.
Methods
Patient history
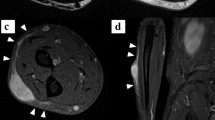
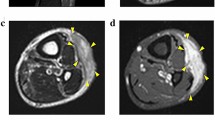
A 94-year-old Caucasian woman presented in May 2014, at the Department of Orthopedic Surgery at the Medical University of Graz with a soft tissue lump. Radiography and magnetic resonance imaging (MRI) revealed a tumour mass with an extent of 5 × 3.5 × 4.5 cm on the left knee. After a biopsy a wide resection was performed at our department and the knee joint was covered with a gastrocnemius muscle flap. The postoperative histopathological report revealed a MFS G3 with resection margins free of disease. The patient received adjuvant radiotherapy. The research reported in this study was conducted adhering to the principles of human welfare according to the Consort declaration on clinical research design and the Helsinki declaration on medical protocols and ethics. The study protocol and the informed consent of the patients were approved by the ethics committee of the Medical University Graz (vote #27–258ex14/15; valid until 17.04.2016). The patient was extensively informed and gave her written approval.
Cell culture procedures
The tumour tissue was obtained immediately after surgical removal. After mechanical disaggregation of the tumour tissue into 1–2 mm3 pieces, the minced tissue was enzymatically digested with 2 mg/ml collagenase B (Roche Diagnostics, Mannheim, Germany) for approximately 20 h under constant rotation at 37˚C. After centrifugation at 1400 rpm for 5 min two fractions were isolated. On the one hand, the cell pellet was washed twice with PBS and plated in Dulbecco’s-modified Eagle’s medium (DMEM-F12; Invitrogen, Darmstadt, Germany), containing 10% foetal bovine serum (FBS), 1% L-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin and 0.25 μg amphotericin B (all Invitrogen). From this fraction the sub-clone MUG-Myx2b was grown. On the other hand, the viscous colloidal supernatant was collected and cultured as sub-clone MUG-Myx2a in the above-mentioned culture medium. During cultivation time, the cells were regularly cryopreserved. Cells grew to be adherent as a monolayer and were passaged more than 70 times during the period of 24 months. Even from the beginning of cultivation MUG-Myx2a and MUG-Myx2b differed regarding their morphology and growth behaviour (see Fig. 1). All cell cultures were periodically checked for mycoplasma by PCR.
Cell line identification Power Plex® 16 System
Frozen tumour tissue from the patient was dissected into small pieces and re-suspended in 180 μl ATL buffer (Qiagen, Hilden, Germany). Cell pellets (5 × 105) from both clones (passage 45 and passage 48) were re-suspended in 200 μl PBS; subsequently 20 μl Proteinase K and 200 μl AL Buffer (Qiagen) were added. DNA preparations were performed using the QIAamp DNA Mini kit (Qiagen) in accordance with the manufacturer’s protocol. After normalising the DNA, 1 ng DNA of each sample was amplified using the Power Plex® 16 System (Promega, Vienna, Austria) in a 10 μl reaction. One μl of the product was mixed with Hi-Di formamide (Applied Biosystems Inc., Foster City, US) and Internal Lane Standard (ILS600), denatured and fractionated on an ABI 3730 Genetic Analyser (Applied Biosystems Inc.). The resulting data were processed and evaluated using ABI Genemapper 4.0 (Applied Biosystems Inc.).
Immunohistochemical (IHC) and immunofluorescence (IF) stainings
Patient’s tumour
For the histopathological evaluation, the tumour was tested using the streptavidin-biotin peroxidase complex method with antibodies against Caldesmon (Dako, Glostrup, Denmark), S100 (Dako), CD34 (Neomarkers, Fremont, CA), Desmin, EMA, and Pan-CK (all Ventana Medical Systems, Tucson, AZ).
MUG-Myx2a/b morphology
For IF analysis, cells were seeded at a concentration of 1 × 104 cells on polystyrene culture slides (BD Biosciences, San Diego, CA). Slides were washed with PBS and fixed by exposure to formalin 4% for 10 minutes. After drying and rehydration, the slides were treated with Large Volume UltraV-Block (ThermoScientific, Waltham, US) for 10 min at room temperature to block nonspecific binding, incubated with the primary monoclonal mouse anti-Vimentin antibody (Dako) for 30 min and, after several washing steps, incubated with the Cy2 conjugated sheep anti-mouse IgG secondary antibody (Jackson Immunoresearch, Suffolk, UK) at a dilution of 1:200 for 30 min. Nuclei were counterstained with DAPI (Invitrogen).
Mice tissue
IHC studies using the streptavidin-biotin peroxidase complex method were carried out on histological slides from MUG-Myx2a and MUG-Myx2b mice tumours, employing a rabbit monoclonal primary antibody against the anti-Ki-67 (clone 30–9) (Ventana Medical Systems) using the BenchMark Ultra instrument (Ventana Medical Systems). Slides were photographed using an Olympus BX51 microscope with an Olympus DP71 microscope digital camera. The stained slides were scanned digitally and positive and negative cells were quantified using the ImageScope software (ImageScope Virtual Slide, version 6.25, Aperio Technol., Vista, US). Positivity was determined by assessing the number of positive cells / number of total cells.
Cell proliferation analysis
xCELLigence System
The xCELLigence RTCA-DP device from Roche Diagnostics (OLS, Bremen, Germany) was used to monitor cell proliferation in real-time. Respectively 5 × 103 MUG-Myx2a/b cells were seeded in electronic microtiter plates (E-Plate™, OLS) and measured for 72 h with the xCELLigence system according to the instructions in the user’s manual. Cell density measurements were performed in quadruplicate with a programmed signal detection every 20 min. Data acquisition and analyses were performed with the RTCA software (version 1.2, OLS).
Growth behaviour
2.5 × 103 MUG-Myx2a/b cells were seeded into 96-well microtiter plates (Brand, Voerde-Friedrichsfeld, Germany) and the CellTiter 96® AQueous Assay (Promega, Mannheim, Germany) was performed after the manufacturer’s instructions at days 1, 2, 3, and 6. The culture medium was used as a negative control.
Drug sensitivity assay
MUG-Myx2a and MUG-Myx2b were adjusted to a density of 3 × 103 cells and incubated in 96-well microplates. The cells were exposed to various concentrations of chemotherapeutic drugs (the DNA topoisomerase II inhibitor doxorubicin hydrochloride, the histone deacetylase inhibitors inhibitor Verinostat, and the proteasome inhibitor Bortezomib; Selleckchem, Houston, TX) for 48 h. Drug sensitivity was determined by the MTS assay following the manufacturers’ instructions in quadruplicate using a photometer (Spektramax; BMG Labtech., Offenburg, Germany) at the wavelength of 490 nm. Results were expressed as the mean from three independent experiments (n = 3, measured in biological quadruplicates) and error bars represent the S.D.
Real-time chemotaxis assay
Chemotaxis assays were performed in cell invasion and migration (CIM-Plate™; OLS) using an xCELLigence RTCA-DP instrument (OLS). A CIM-plate comprises two separate parts: an upper chamber with a microporous membrane (pore size = 8 mm) embedded with gold microelectrodes on its underside and a lower chamber. This migratory assay is based on a small and harmless electrical current applied between the electrodes. Cell adherence to this surface while migrating from the upper chamber toward 10% FBS as chemoattractant placed in the lower chamber is measured. The more cells accumulate on the underside of the membrane, the more the electrical current is impeded by the insulating properties of cellular plasma membranes. The overall cell-induced impedance is measured in real-time as the cell index (CI), a dimensionless parameter.
Tumour formation
Tumourigenicity of MUG-Myx2a/b clones: 7 week old female nu/nu Foxn1 mice (Charles River Laboratories, Sulzfeld, Germany) were xenotransplanted with MUG-Myx2a (p49) and MUG-Myx2b (p52) cells. Respectively 5 × 106 cells were suspended in 0.2 ml of serum-free medium and subcutaneously inoculated into the right and left flank of 5 mice. The mice were observed daily and the tumour growth was monitored. All animal work was done in accordance with a protocol approved by the institutional animal care and use committee at the Austrian Federal Ministry for Science and Research (BMWF) (vote 66.010/0160-II/3b/2012).
Mutation Analysis by Next-Generation Sequencing (NGS)
Genomic DNA of MUG-Myx2a and MUG-Myx2b was isolated on a Maxwell, MDxResearch System (Promega). Highly multiplexed PCR was used to generate amplicon libraries covering the coding region of 50 genes commonly implicated in cancer (Cancer Hotspot Panel v2, Cat.Nr. 4475346; Thermo Fisher Scientific, Waltham, MA). All analyses were performed in duplicates. Libraries were prepared using the Ion AmpliSeq Library Kit 2.0 (Cat.Nr. 4475345; Thermo Fisher Scientific) and sequencing was performed on an Ion Proton Sequencer (Thermo Fisher Scientific). Emulsion PCR and sequencing runs were performed with the appropriate kits (Ion One Touch Template Kit version 2 and Ion Proton 200 Sequencing Kit; Thermo Fisher Scientific) using Ion PI chips. Sequencing length was set to 520 flows and yielded reads ranging from 70 to 150 bp, consistent with the expected amplicon size range.
NGS Data Analysis
Initial data analysis was performed using the Ion Torrent Suite Software version 4.1 Plug-ins (Thermo Fisher Scientific, open source, general public license). Briefly, this included base calling, alignment to the reference genome (HG19) using the Torrent Mapper and variant calling by a modified diBayes approach by taking into account the flow space information. All called variants were annotated using open source software (Annovar34, http://www.openbioinformatics.org/annovar/annovar download form.php, and SnpEff35, http://snpeff.sourceforge.net/download.html; both last accessed September 28, 2015). Coding, nonsynonymous mutation calls present in both duplicate analyses were further evaluated and visually inspected in Integrative Genomics Viewer. Variant calls resulting from technical read errors or sequence effects were excluded from the analysis. Mutations were subsequently assessed for biological relevance by computerized prediction (in silico prediction). Sorting Intolerant From Tolerant algorithm (SIFT) was used to generate SIFT scores.
Low density whole genome copy number variation (CNV)
NGS libraries werer prepared using 500 ng genomic DNA and the Ion Plus Fragment Library Kit (Cat.Nr. 4471252, Thermo Fisher Scientific). DNA libraries were sequenced on the Ion Proton Sequencer yielding 8–10 million reads per library. CNV analysis was performed using the CNAnorm Bioconductor R package36. Briefly, reads were aligned to hg19 and summarized in windows of 10 kB size. CNAnorm was used to normalize for GC content and perform CNV analysis using coverage data from a non neoplastic reference sample. Circos plots were created using the circos software package37 (http://circos.ca/). Ploidy analysis of single genes was calculated in R using gene coordinates from ensembl database and ploidy ratio information from CNAnorm.
Statistical analysis
The outcome variables were expressed as mean ± SD. The student’s unpaired t-test and the exact Wilcoxon’s test were used to evaluate differences between groups with the PASW statistics 18 software (IBM Corporation, Somers, NY). Two-tailed P-values below 0.05 were considered statistically significant. Graphic data were prepared with SigmaPlot® (Systat Software Inc., San Jose, US).
Additional Information
How to cite this article: Lohberger, B. et al. Establishment of a novel cellular model for myxofibrosarcoma heterogeneity. Sci. Rep. 7, 44700; doi: 10.1038/srep44700 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Fletcher, C. D. M., Hogendoorn, P. C. W. & Mertens, F. In World Health Organization Classification of Tumours of Soft Tissue and Bone 4th edn, Ch. 15, 264–274 (IARC Press, 2013).
Damron, T. A., Ward, W. G. & Stewart, A. Osteosarcoma, chondrosarcoma, and Ewing’s sarcoma. National Cancer Data Base Report. Clin. Orthop. Relat. Res. 459, 40–47 (2007).
Giuffrida, A. Y. et al. Chondrosarcoma in the United States (1973 to 2003): an analysis of 2890 cases from the SEER database. J. Bone Joint Surg. Am. 91 (5), 1063–1072 (2009).
Willems, S. M. et al. Cellular/intramuscular myxoma and grade I myxofibrosarcoma are characterized by distinct genetic alterations and specific composition of their extracellular matrix. J. Cell Mol. Med. 13, 1291–1301 (2009).
Willems, S. M. et al. Imaging mass spectrometry of myxoid sarcomas identifies proteins and lipids specific to tumour type and grade, and reveals biochemical intratumour heterogeneity. J. Pathol. 222 (4), 400–409 (2010).
Jones, E. A. et al. Multiple statistical analysis techniques corroborate intratumor heterogeneity in imaging mass spectrometry datasets of myxofibrosarcoma. PLoS One 6 (9), e24913 (2011).
Burrell, R. A., McGranahan, N., Bartek, J. & Swanton, C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 501, 338–345 (2013).
Burrell, R. A. & Swanton, C. Tumour heterogeneity and the evolution of polyclonal drug resistance. Mol. Oncol. 8 (6), 1095–1111 (2014).
Marusyk, A., Almendro, V. & Polyak, K. Intra-tumour heterogeneity: a looking glass for cancer? Nat. Rev. Cancer 12, 323–334 (2012).
Holohan, C., Van Schaeybroeck, S., Longley, D. B. & Johnston, P. G. Cancer drug resistance: an evolving paradigm. Nat. Rev. Cancer 13, 714–726 (2013).
Swanton, C. Intratumor heterogeneity: evolution through space and time. Cancer Res. 72, 4875–4882 (2012).
Meacham, C. E. & Morrison, S. J. Tumour heterogeneity and cancer cell plasticity. Nature 501, 328–337 (2013).
Gerlinger, M. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892 (2012).
Sottoriva, A. et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc. Natl. Acad. Sci. USA 110, 4009–4014 (2013).
Conway, C. et al. Next-generation sequencing for simultaneous determination of human papillomavirus load, subtype, and associated genomic copy number changes in tumors. J. Mol. Diagn. 14 (2), 104–111 (2012).
Snijders, A. M. et al. Assembly of microarrays for genome-wide measurement of DNA copy number. Nat. Genet. 29, 263–264 (2001).
Nielsen, G. P., Burns, K. L., Rosenberg, A. E. & Louis, D. N. CDKN2A gene deletions and loss of p16 expression occur in osteosarcomas that lack RB alterations. Am. J. Pathol. 153 (1), 159–163 (1998).
Levine, A. J. p53, the cellular gatekeeper for growth and division. Cell 88, 323–331 (1997).
Latres, E. et al. Chromosome 17 abnormalities and TP53 mutations in adult soft tissue sarcomas. Am. J. Pathol. 145, 345–355 (1994).
Myers, M. P. et al. PTEN, the tumor suppressor from human chromosome 10q23, is a dual-specificity phosphatase. Proc. Natl. Acad. Sci. USA 94, 9052–9057 (1997).
Li, J. et al. PTEN, a putative protein tyrosinephosphatase gene mutated in human brain, breast, and prostate cancer. Science 275, 1943–1947 (1997).
Stambolic, V. et al. Regulation of PTEN transcription by p53. Mol. Cell 8, 317–325 (2001).
Chang, C. J. et al. PTEN nuclear localization is regulated by oxidative stress and mediates p53-dependent tumor suppression. Mol. Cell Biol. 28, 3281–3289 (2008).
Puzio-Kuter, A. M. et al. Inactivation of p53 and PTEN promotes invasive bladder cancer. Genes Dev. 23, 675–680 (2009).
Pollock, R. et al. Wild-type p53 and a p53 temperature-sensitive mutant suppress human soft tissue sarcoma by enhancing cell cycle control. Clin. Cancer Res. 4, 1985–1994 (1998).
Zhang, L. et al. Wild-type p53 suppresses angiogenesis in human leiomyosarcoma and synovial sarcoma by transcriptional suppression of vascular endothelial growth factor expression. Cancer Res. 60, 3655–3661 (2000).
Yin, L. et al. Mutational analysis of p53 and PTEN in soft tissue sarcoma. Mol. Med. Rep. 5 (2), 457–461 (2011).
Wen-Hann, T. et al. The spectrum of vascular anomalies in patients with PTEN mutations: implications for diagnosis and management. J. Med. Genet. 44, 594–602 (2007).
Rodríguez-Escudero, I. et al. A comprehensive functional analysis of PTEN mutations: implications in tumor- and autism-related syndromes. Hum. Mol. Genet. 20 (21), 4132–4142 (2011).
Wang, Y. et al. Polymorphisms of KDR gene are associated with coronary heart disease. J. Am. Coll. Cardiol. 50: 760–767 (2007).
Zhang, W. et al. VEGF receptor-2 variants are associated with susceptibility to stroke and recurrence. Stroke 40, 2720–2726 (2009).
Lasota, J. & Miettinen, M. KIT and PDGFRA mutations in gastrointestinal stromal tumors (GISTs). Semin. Diagn. Pathol. 23 (2), 91–102 (2006).
Hernandez, S. et al. FGFR3 mutations in prostate cancer: association with low-grade tumors. Mod. Pathol. 22, 848–856 (2009).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from next-generation sequencing data. Nucleic Acids Res. 38 (16), e164 (2010).
Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6 (2), 80–92 (2012).
Gusnanto, A., Wood, H. M., Pawitan, Y., Rabbitts, P. & Berri, S. Correcting for cancer genome size and tumour cell content enables better estimation of copy number alterations from next-generation sequence data. Bioinformatics 28 (1), 40–47 (2012).
Krzywinski, M. et al. Circos: an Information Aesthetic for Comparative Genomics. Genome Res. 19, 1639–1645 (2009).
Acknowledgements
The authors would like to thank Heike Kaltenegger, Andrea Thüringer, Iris Halbwedl, and Elke Winter for their technical assistance, and Nicholas Donohue for English-language proofreading. Financial support from the Medical University of Graz is gratefully acknowledged.
Author information
Authors and Affiliations
Contributions
B.Lo. conceived and supervised the study. N.S., H.K., and K.K. performed the experiments. B.Lo., S.S., B.R. and K.K. analysed and interpreted the data. B.Lo., K.K., B.Li., and A.L. drafted and revised the manuscript. B.Li. and A.L. provided technical support. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Lohberger, B., Stuendl, N., Leithner, A. et al. Establishment of a novel cellular model for myxofibrosarcoma heterogeneity. Sci Rep 7, 44700 (2017). https://doi.org/10.1038/srep44700
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep44700
- Springer Nature Limited
This article is cited by
-
Clear surgical margins as a prognostic indicator for disease recurrence, with no impact on survival rates in patients with myxofibrosarcoma
Scientific Reports (2024)
-
A novel patient-derived immortalised cell line of myxofibrosarcoma: a tool for preclinical drugs testing and the generation of near-patient models
BMC Cancer (2023)
-
Molecular and clinicopathological analysis revealed an immuno-checkpoint inhibitor as a potential therapeutic target in a subset of high-grade myxofibrosarcoma
Virchows Archiv (2022)
-
Establishment and characterization of NCC-MFS3-C1: a novel patient-derived cell line of myxofibrosarcoma
Human Cell (2021)
-
Establishment and characterization of NCC-MFS2-C1: a novel patient-derived cancer cell line of myxofibrosarcoma
Human Cell (2021)