Abstract
Molecular graph representation learning has shown considerable strength in molecular analysis and drug discovery. Due to the difficulty of obtaining molecular property labels, pre-training models based on self-supervised learning has become increasingly popular in molecular representation learning. Notably, Graph Neural Networks (GNN) are employed as the backbones to encode implicit representations of molecules in most existing works. However, vanilla GNN encoders ignore chemical structural information and functions implied in molecular motifs, and obtaining the graph-level representation via the READOUT function hinders the interaction of graph and node representations. In this paper, we propose Hierarchical Molecular Graph Self-supervised Learning (HiMol), which introduces a pre-training framework to learn molecule representation for property prediction. First, we present a Hierarchical Molecular Graph Neural Network (HMGNN), which encodes motif structure and extracts node-motif-graph hierarchical molecular representations. Then, we introduce Multi-level Self-supervised Pre-training (MSP), in which corresponding multi-level generative and predictive tasks are designed as self-supervised signals of HiMol model. Finally, superior molecular property prediction results on both classification and regression tasks demonstrate the effectiveness of HiMol. Moreover, the visualization performance in the downstream dataset shows that the molecule representations learned by HiMol can capture chemical semantic information and properties.
Similar content being viewed by others
Introduction
In recent years, molecular representation learning has attracted growing attention in the study of chemical drug analysis and discovery1. Motivated by the remarkable success of machine learning2, especially deep learning3, molecular representation learning has increasingly explored machine learning-based methods. These methods are widely used for various molecular applications, such as chemical property prediction4,5, drug molecule generation6,7,8, and optimization9. How to learn comprehensive and effective molecular representation remains an open challenge.
Inspired by the development of Natural Language Processing (NLP)10,11,12, a range of methods apply language models to handle string-based molecular representations like SMILES13 and SELFIES14. Specifically, these molecular strings are encoded by Recurrent Neural Network (like Gated Recurrent Unit (GRU)15 and Long Short-Term Memory (LSTM)16 or Transformer12, which can be trained in a supervised manner by predicting molecular properties17. Further, to handle enormous unlabeled data, many works pay attention to self-supervised learning (SSL) frameworks. The SSL decoders generally generate the original strings18 or other random SMILES of input molecules19 or recover masked tokens of molecular strings20. Due to the single dimension of strings, important topology structures of molecular graphs are ignored. Therefore, many researchers have transformed their interests from 1D molecular strings to 2D graphs21,22. In view of the success of SSL, graph SSL-based pre-training frameworks23,24,25,26,27 have been increasing rapidly in the past few years. They capture the topology of 2D graphs and have also shown a positive effect in molecular analysis tasks. However, their commonality for all kinds of graphs leads to the neglect of unique structural properties of chemical molecules, such as rings and functional groups.
Many recent presentation learning works5,22,28,29,30,31,32,33 consider the characteristic of molecular graphs. Zhang et al.32 model a clustering problem to learn molecular motifs, and the GNN-encoded atom representations are grouped into subgraphs for contrastive learning. Zhang et al.30 design self-supervised molecular pre-training framework, whose pretext task is predicting the motifs based on a given order (depth-first search or breadth-first search) on the graph. Wang et al.5 present three molecular graph structural augmentation patterns and aligns different augmentations of the same molecule through contrastive learning. Wang et al.33 also propose a contrastive learning framework for molecular learning, in which not only molecular pairs but also motif pairs are sampled for contrastive pre-training. Wang et al.31 takes into account the chemical reaction relationships between the molecules, i.e., the sum of molecular representations of all reactants is supposed to equal that of all products. Despite the improvement of molecule representation learning, previous works still fail to solve the following problems and challenges: (1) How to preserve and capture molecular structure adequately? Many recent molecular learning methods5,28 apply graph augment to construct different views and contrast multiple views for pre-training. However, some general graph data augmentations (like edge modification34 or graph diffusion35) tend to destroy the structure or attributes of molecules, so some important chemical properties are highly possible to be buried. In addition to the whole topology structure, the motif is also valuable and has an important impact on the molecular properties. Therefore, it is beneficial and challenging for molecular learning to preserve the complete molecular structure and directly incorporate the motifs. (2) How to fuse more comprehensive information into molecular graph representations? With the development of GNN36,37,38,39,40, growing molecular learning methods29,31 leverage GNN as the encoder backbone, in which nodes flow information among local neighbors, and the graph representation is obtained by integrating all its node representations via READOUT function. The pattern of aggregating neighborhood representations lacks a global scale. Moreover, the READOUT operation can only transfer information from low-order atoms to the high-order molecule, but can not implement the interaction between them. (3) How to design the pretext tasks of self-supervised pre-training? Self-supervised pre-training requires design pretext tasks to optimize the backbone model parameters. The pretext tasks affect the transferable ability of the pre-training model to a great extent, which determines whether the model can achieve satisfactory fine-tuning performance in downstream tasks. Therefore, it is key for self-supervised pre-training models to design reasonable pretext tasks to improve transferable ability.
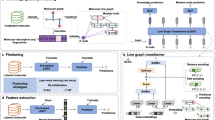
To resolve the aforementioned challenges, we propose a self-supervised learning framework for molecular property prediction called Hierarchical Molecular Graph Self-supervised Learning (HiMol). Figure 1 shows the overall framework of HiMol. HiMol consists of two major components, Hierarchical Molecular Graph Neural network (HMGNN) and Multi-level Self-supervised Pre-training (MSP). HMGNN presents a GNN-based hierarchical molecule encoder, aiming at addressing the first two challenges. To better explore the unique internal chemical structure of the molecular graphs, we improve rules to decompose the molecular motif structure. The molecular motif denotes the substructure of a molecule, which contains many chemical characteristics in general. For instance, the carboxyl group usually indicates acidity. The motif decomposition rules conform to chemical criteria to avoid destroying the chemical characteristics. We add motifs into the molecular graph as nodes to mine implicit semantic information. In addition, we augment a graph-level node to learn molecular graph representation through training along with normal nodes and motifs, in place of the READOUT function. The graph-level node collects information from all nodes and transfers the global information back to nodes through motif-level nodes. This circulation realizes the information interaction between different orders, which helps the model to learn more comprehensive representations. MSP designs multi-level pretext tasks as supervision signals of pre-training. Specifically, MSP designs three generative tasks that predict bond links, atom types, and bond types with the atom representations; MSP designs two predictive tasks that predict the number of atoms and bonds with the molecule representation. Compared with single-signal pre-training frameworks, multi-level pretext tasks help fuse more molecular information into molecule representations from diverse perspectives.
HMGNN: the input molecular graph is first decomposed into motifs, which are constructed as motif-level nodes. Further, a graph-level node is added. The augmented graph is constructed by adding node-motif and motif-graph edges. GNN is employed to learn hierarchical node representations on the augmented graph. MSP: encoded atom representations and molecular graph representation predict five tasks for self-supervised pre-training.
In summary, the advantages of our HiMol are as follows:
-
HiMol builds a hierarchical GNN to encode molecules. It facilitates incorporating multi-order information into the molecule representation. To the best of our knowledge, HiMol is the first molecular representation learning method that encodes node-motif-graph hierarchical information.
-
HiMol constructs motifs based on chemical rules and mine the characteristics of molecular substructures without destroying chemical structures.
-
HiMol augments a graph-level node to simulate the molecular graph representation. It allows the molecular graph representation to participate in the training directly and realizes bidirectional transmission of local and global features.
-
HiMol designs multi-level self-supervised pre-training tasks in accordance with hierarchical representations of molecules. It improves the transferable potential of the pre-training framework and helps learn more informative molecular representations.
-
HiMol outperforms the state-of-the-arts (SOTA) in the downstream molecular property prediction tasks including classification and regression. It demonstrates the effectiveness of our proposed HiMol.
Results
HiMol framework
Our HiMol presents a self-supervised pre-training framework to encode hierarchical representations of the molecular graph. HiMol mainly includes the Hierarchical Molecular Graph Neural network (HMGNN) and Multi-level Self-supervised Pre-training (MSP).
Given the SMILES of a molecule, we first transform it into a graph \(G=\left(V,E\right)\) based on RDKIT41, where V and E denote atom and bond sets respectively. Then we decompose G into a series of motifs \({V}_{{{{{\rm{m}}}}}}=\left\{{V}_{{{{{\rm{m}}}}}}^{1},{V}_{{{{{\rm{m}}}}}}^{2},...,{V}_{{{{{\rm{m}}}}}}^{k}\right\}\), k is the number of motifs. The decomposition of motifs adds a rule on the basis of BRICS42, i.e., break large ring fragments and select their minimum rings as generated motifs. All motif nodes Vm are incorporated into V, and edges between the corresponding atoms and motifs Em are linked and merged into E. In addition, we augment a graph-level node Vg, meanwhile, edges between all motifs and Vg are appended into the initial graph G. The augmented graph \(\widetilde{G}\) is input into GNNs to learn the hierarchical presentations, including atom-level representations \({{{{{{{{\bf{H}}}}}}}}}_{{{{{{{{\rm{a}}}}}}}}}\in {{\mathbb{R}}}^{|V| \times d}\), motif-level representations \({{{{{{{{\bf{H}}}}}}}}}_{{{{{{{{\rm{m}}}}}}}}}\in {{\mathbb{R}}}^{k\times d}\), and graph-level representation \({{{{{{{{\bf{H}}}}}}}}}_{{{{{{{{\rm{g}}}}}}}}}\in {{\mathbb{R}}}^{d}\). In the process of MSP, the atom-level representations are used to predict atom types, bond links, and bond types; the graph-level representation is used to predict the number of atoms and bonds. Cross entropy loss and smooth L1 loss are respectively applied to optimize the atom- and molecule-level learning.
As for the fine-tuning for downstream tasks, the graph-level representations pass through a 2-layer Multi-Layer Perceptron (MLP) to predict molecular properties. The pre-trained GNN weights are transferred to the fine-tuning model and continue to be updated along with the MLP parameters under the supervision of labels. More details about HiMol method are shown in “Methods”.
Molecular property prediction
To evaluate the effectiveness of HiMol for molecular property prediction, we conduct classification and regression tasks on diverse datasets from MoleculeNet. Table 1 reports the mean and standard deviation of ROC-AUC (%) results on the binary classification tasks. We summarize the observations from the results as follows. (1) Our HiMol achieves the strongest performance on four out of six datasets. For the remaining datasets, HiMol still reaches competitive performance on Tox21. Although MolCLR shows remarkable results on ClinTox, its performance on other datasets is not satisfactory. On average, HiMol improves by 2.4% over the best-performing baseline. The enhancement proves the superiority and effectiveness of our hierarchical graph learning model HiMol. (2) Among all SOTA methods, two motif-based models G_Motif and MGSSL outperform other baselines, which indicates the capture of motif structure play an important role in molecular graph learning. However, they only use motif prediction as the pretext task of pre-training, the hidden representations extracted by GNNs do not directly incorporate motif information during the fine-tuning for the downstream tasks. Our HiMol encodes motif structures through the GNN backbone, which benefits the molecular representation to better learn and predict the internal properties of molecules during fine-tuning. (3) We give two different scales of HiMol models, in which the layer numbers of GIN for HiMol (SMALL) and HiMol (LARGE) are respectively 3 and 5. The LARGE version performs slightly better, which may be explained as the deeper GNN being able to capture more structural information.
The regression results are shown in Table 2. According to the recommendation of MoleculeNet, mean-absolute error (MAE) is used as an evaluation metric for physical chemistry datasets (ESOL, FreeSlov, and Lipophilicity) while root-mean-square error (RMSE) is the metric for quantum mechanics datasets (QM7, QM8, and QM9). The observations of Table 2 are summarized as follows. The regression result of MoCL28 on QM9 is not given since it is expensive to calculate the similarity matrix in the process of pre-training on QM9, and no corresponding result is given in the official paper. (1) HiMol outperforms all baselines on five out of six datasets and reaches competitive performance on the remaining QM8. Notably, the MAE decreases by 55.5% over the strongest baseline on the challenging dataset QM9. (2) Similar to the results of classification tasks, the LARGE version has a small edge over the SMALL version in the regression tasks.
Visualization of molecular representations
To exhibit intuitively the learned representations by HiMol, we utilize t-SNE43 to project them to a two-dimensional space and different colors to distinguish molecular property labels. Note that we simply visualize the fine-tuned molecular presentations in the test set, which do not incorporate groundtruth. From Fig. 2, molecular presentations are grouped in line with their labels on both classification and regression tasks. For example, in Fig. 2a, the Class label represents the binary results of binding for BACE-1 inhibitors. Molecules labeled 0 and 1 are grouped on the top left and bottom right, respectively. Similarly, in Fig. 2b, molecules have increasingly high exp values from the top left to bottom right. Moreover, in Fig. 2c, d, it can be observed that the visualized presentations have a high degree of consistency in terms of two important properties of molecules on QM9. The visualization in Fig. 2 demonstrates that our HiMol can extract the internal properties of molecules. Similar conclusions can be observed in other datasets (see Supplementary Fig. 1).
a BACE (Class): color represents binary labels of binding results for BACE-1 inhibitors. b Lipophilicity (exp): color represents octanol/water distribution coefficient. c QM9 (homo): color represents highest occupied molecular orbital energy (homo) of molecules. d QM9 (lumo): color represents lowest unoccupied molecular orbital energy (lumo) of molecules.
To further demonstrate the effectiveness of our pre-training framework, we visualize the molecular representations fine-tuned in downstream tasks without pre-training in Supplementary Fig. 2. Compared with Fig. 2, the visualization results without pre-training are chaotic in terms of molecules with different properties. The pre-training improves the performance of the HMGNN encoder and is beneficial to learning more chemical properties in downstream tasks.
Molecular representation similarity ranking
To further investigate the semantic information contained in the pre-trained molecular representations via HiMol, we evaluate the similarity between the query molecule and other molecules and draw the top-five most similar molecules. The cosine similarity between the query molecule q and the candidate molecule c is calculated as follows:
where \({{{{{{{{\bf{H}}}}}}}}}_{{{{{{{{\rm{g}}}}}}}}}^{q}\in {{\mathbb{R}}}^{d}\) and \({{{{{{{{\bf{H}}}}}}}}}_{{{{{{{{\rm{g}}}}}}}}}^{c}\in {{\mathbb{R}}}^{d}\) are the graph representations of the query molecule and candidate molecule, respectively. Then the similarity of all candidate molecules is ranked and the top-five molecules of the query ZINC9452931 are displayed in Fig. 3. From the figure, we can observe that the five molecules have similar structures and functional groups with the query. Specifically, the top-five similar molecules possess all atom types of the query, i.e., C, H, N, O, S. Moreover, the structure that two rings share a common edge exists in all molecules. Top@1 molecule not only have similar ring structures with the query, but also the same chain structures like NC(=O) and C(=O). The molecular representation similarity ranking experiments imply that our framework HiMol can learn molecular representation with chemical semantic information. Meanwhile, encoding the motifs is conducive to identifying molecular substructures like functional groups and rings. More results about other query molecules are shown in Supplementary Fig. 3.
Fine-tuning mode analysis
To investigate the impact of different fine-tuning strategies on the model prediction tasks, we explore freezing mode and full fine-tuning mode. They both inherit the pre-trained weights of the hierarchical molecular graph neural network. The freezing mode freezes the parameters of HMGNN and only updates the weights of downstream classifiers, while the full fine-tuning mode trains all weights of HMGNN and classifiers in an end-to-end manner during fine-tuning. In addition, we evaluate HiMol without pre-training and vanilla GIN38, which are directly trained through supervised downstream tasks. Table 3 shows two different versions of all methods, where SMALL version and LARGE version encode molecular graphs by 3-layer and 5-layer GNNs, respectively. From the results, we can observe that: (1) Full fine-tuning mode presents more advantage over freeze mode, which indicates that our proposed HMGNN plays a decisive role for prediction tasks; (2) Himol without pre-training exhibits better performance to GIN, which demonstrates that HMGNN has the ability to extract more abundant structural information; (3) HiMol achieves better performance after pre-training, which verifies that our proposed multi-level self-supervised pre-training strategy can effectively train model parameters and transfer knowledge to downstream data.
Figure 4 further displays the train and test classification ROC-AUC curves of the four aforementioned modes. It can be observed that the full HiMol can converge faster to optimum performance than other methods. Besides, the pre-trained methods (HiMol and its freeze mode) have better stability than the other two modes without pre-training in terms of training and testing curves, indicating that our pre-training framework boosts the stability of fine-tuning.
Ablation studies on pre-training
To analyze the effectiveness of each part of our proposed HiMol framework, we conduct ablation experiments on HMGNN and MSP during the pre-training process. Figure 5 illustrates the ablation experimental results of HMGNN and MSP.
In Fig. 5a, we compare HiMol with two pre-training versions. HMGNN w/o motif-level denotes that molecular graphs only augment graph-level nodes and node-graph edges, without decomposing motifs. HMGNN w/o graph-level obtains graph-level representations through performing mean pooling to motif-level presentations. We can observe the following insights: (1) The performance of HiMol removing motifs becomes worse on all benchmarks, which implies the important role of encoding motifs for molecular representation learning. HiMol decomposes motifs to learn hierarchical representations, which is beneficial to learn more molecular structural characteristics and functions. (2) Replacing the graph-level node representation with the squeezed representation via graph pooling also makes the molecular property prediction performance slightly decrease. Augmenting a graph-level node is superior to the mean-pooling function, since it can indirectly learn multi-order features through the motif node, and can transfer the global information to all atom nodes during training. Mutually, atom nodes not only integrate local neighbor information but also capture substructural and global graph information. These multi-level interactions between normal nodes and the graph-level node improve the learning performance of HMGNN.
The ablation versions for MSP are shown in Fig. 5b. MSP w/o atom-level contains only graph-level pretext tasks in the pre-training (the number of atoms and bonds). Similarly, MSP w/o graph-level contains only atom-level pretext tasks (bond links, atom types, and bond types). MSP w/o α simply sums all the loss values, removing the α weight parameter to balance. It can be observed that the combination of multi-level self-supervised tasks promotes the comprehensiveness transfer ability of pre-training compared with a single pretext task. In addition, the learnable weight α adjusts the importance of the loss of different parts, which can better coordinate the relationship between multi-level tasks to improve the performance of pre-training.
Discussion
Motif encoding
First of all, we propose to encode the constructed motifs while learning the atom representations. HiMol mines the substructure of molecules through motifs. There are many existing molecular representation learning methods29,30 considering motif structure. They generally decompose molecules in a large-scale database into motifs and build a motif dictionary, then motif prediction serves as self-supervised pretext tasks in the pre-training process of molecular representation learning. However, these methods have some disadvantages: (1) When the constructed motif dictionary is large, the computation of motif prediction is expensive. (2) Motifs as the self-supervised signal only, are difficult to be incorporated by hidden representations directly. We encode motifs contained in each molecule in the encoder part, which directly integrates the motif structure into the hidden representations. Moreover, the augmentation of motif-level nodes better captures the affiliation relationship between atoms and motifs.
Graph encoding
In addition to motif-level nodes, we also augment a graph-level node into the molecular graph, which participates in the aggregation process to extract molecule representation. Most molecular learning methods use vanilla GNNs as the backbones and squeeze node representations into graph representation through the READOUT function, i.e., graph pooling methods like MAX, SUM, and MEAN. The process of GNNs is introduced in Supplementary Note 1. It is convenient to handle varying sizes of graph data, but the graph-level representation fails to be involved in learning. BERT model11,44 proposes to attach a token to the sequence, whose representation is regarded as the sequence-level feature. Referring to BERT, Graphormer45 adds a virtual node into the graph, which is linked and updated with all nodes, and the feature of the virtual node is the graph representation. Inspired by the success of the aforementioned works, we augment a graph-level node to extract the molecular graph representation via participating in aggregation and updating with all atoms. Nevertheless, related methods only connect the graph-level node with normal nodes, they are not conducive to capturing subgraph structure. To integrate motif nodes, we connect graph-level nodes with motif nodes to achieve hierarchical aggregation. Compared with the READOUT, our HiMol implements the training and updating of graph nodes and realizes the interaction with multi-order nodes after incorporating molecular motif structure.
Conclusion
In this paper, we propose Hierarchical Molecular Graph Self-supervised Learning (HiMol) to learn molecular presentations. Specifically, we design Hierarchical Molecular Graph Neural Network (HMGNN) backbone to encode node-motif-graph hierarchical representations, which mines chemical semantic information implied at motifs and realizes the interaction of multi-order features. Furthermore, we build multi-level molecular-dependent tasks as self-supervised signals, capturing more comprehensive information from multiple perspectives. Extensive molecular property prediction experiments demonstrate that HiMol shows great improvements over SOTA methods.
The superior performance of molecular representations reflects the potential of our HiMol framework, and there are interesting and promising future works on the basis of HiMol. (1) Combining molecular features with different dimensions, in addition to 2D graphs. (2) Simulating the generative process of molecules and extending our model to molecule generation and optimization tasks.
Methods
Hierarchical Molecular Graph Neural network
We propose a Hierarchical Molecular Graph Neural network (HMGNN) to encode and represent molecular graphs, which mainly contains three parts: (1) motif construction; (2) augmented graph construction; (3) hierarchical representation encoder.
First, we decompose each input molecule graph into a group of motifs. BRICS algorithm is a traditional approach to fragment molecule graphs, which designs chemical rules to break bonds. Specifically, BRICS structurally decomposes molecules into multiple functional fragments according to whether bonds can be combined. However, BRICS splits molecules based on limited chemical rules, resulting in coarse granularity of decomposition. After BRICS decomposition, some large motifs still contain a few more general and functional substructures to be partitioned. Therefore, we add a decomposition rule on the basis of BRICS, i.e., decompose more than one ring substructures connected by common chemical bonds into minimum rings. Figure 6 illustrates the process of motif construction.
All obtained motifs are added as nodes Vm to the molecular graph. Naturally, node-motif edges Em are added between each motif node and all the atom nodes it covers. Since the final output is the molecular graph representation, we construct a graph-level node Vg into the molecular graph and link it to all motif nodes to form motif-graph edges Eg. The final augmented graph is represented as:
On the constructed graph \(\widetilde{G}\), we employ GNN to encode hierarchical molecular representations. Graph Isomorphism Network (GIN)38 is the most widely used to encode molecular graph representation. Inspired by this, we apply GIN as the backbone model of our HiMol, the aggregation pattern of lth layer is given as follows:
where \({{{{{{{{\rm{MLP}}}}}}}}}_{{{{{{{{\rm{a}}}}}}}}}^{l}\) and \({{{{{{{{\rm{MLP}}}}}}}}}_{{{{{{{{\rm{b}}}}}}}}}^{l}\) represent MLP for atom and bond feature transitions atl-layer, respectively, \({{{{{{{{\bf{X}}}}}}}}}_{uv}^{{{{{{{{\rm{b}}}}}}}}}\) denotes the bond feature of uv, and \({{{{{{{{\bf{h}}}}}}}}}_{v}^{0}={{{{{{{{\bf{X}}}}}}}}}_{v}^{{{{{{{{\rm{a}}}}}}}}}\) is the input atom feature of v. The detailed input features of atoms and bonds are shown in Supplementary Table 1. Different from other baselines, the initial molecular features of our HiMol contain degrees of atom nodes and whether bonds are in rings, which contain more molecular structural information. Our proposed HMGNN can encode molecular graphs to obtain multi-level representations simultaneously, including atom-level, motif-level, and molecule-level representations. The learned molecule-level representation can be directly used for downstream tasks, without the READOUT operation.
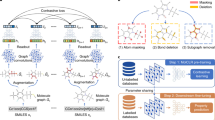
Multi-level Self-supervised Pre-training
After encoding the hierarchical representations through HMGNN, we design generative and predictive pretext tasks to perform Multi-level Self-supervised Pre-training (MSP). The types of self-supervised learning corresponding to different pretext tasks are described in Supplementary Note 2.
For atom-level representations, we design three generative tasks including bond links, atom types, and bond types, aiming at reconstructing graph structure and attributes. Specifically, three 2-layer MLPs with ReLU activation function project atom-level representations to predict three generative tasks, respectively. Cross entropy loss is employed to optimize the atom-level representations as follows:
where yij represents the link between vi and vj, yv,k = 1 represents the atom type of node v is k, and ye,k = 1 represents the bond type of edge e is k. Their corresponding \(\hat{y}\) are the predicted values. Katom and Kbond are the number of atom types and bond types.
For graph-level representations, two predictive tasks on the overall properties of molecules are performed, i.e., the number of atoms and bonds. Similarly, two respective 2-layer MLPs with softplus activation function are applied to predict the graph-level tasks. Based on SmoothL1 loss, the loss of predictive tasks are given in Equations (7) and (8).
where ya and yb are the number of atoms and bonds, respectively; \(\hat{y}\) are predicted values. Supplementary Method describes the transformation process from the representation H to the predicted values y. Finally, we add a learnable vector weight \(\alpha =\left[{\alpha }_{1},{\alpha }_{2},{\alpha }_{3},{\alpha }_{4},{\alpha }_{5}\right]\) normalized by softmax function to balance several losses. The overall objective is calculated as follows:
Datasets
For the pre-training of HiMol, we utilize sampled 250K unlabeled molecules from the ZINC1546 dataset. To evaluate the effectiveness of HiMol, we conduct molecular property prediction experiments on 12 datasets from MoleculeNet47, including six classification task datasets and six regression task datasets. All downstream datasets are split into 80/10/10% for train/validation/test through scaffold-split, which is split in accordance with the molecular structures and provides more challenge for prediction tasks than random-split. The statistical data of all datasets are summarized in Supplementary Table 2.
Baselines
To evaluate the effectiveness of our proposed HiMol, different types of self-supervised learning SOTA are conducted as benchmarks. We compare diverse universal graph learning methods, including GraphSAGE39, GPT_GNN48, AttributeMask49, ContextPred49, InfoGraph50, GraphLoG26, GraphCL25, and JOAO51. Furthermore, we compare several methods designed specifically for molecular graphs, i.e., MoCL28, MolCLR5, G_Motif29, and MGSSL30. The detailed description of baselines is demonstrated in Supplementary Note 3, and the SSL type is summarized in Supplementary Table 3.
Experimental configuration
All experiments are conducted on the Linux server with Nvidia GeForce GTX 1080 Ti GPU and Intel(R) Xeon(R) CPU E5-2620 v4 @ 2.10GHz. In the pre-training phase, the HiMol run 100 epochs using the Adam optimizer with a learning rate of 0.001. We train the LARGE and SMALL versions, with 3-layer and 5-layer GIN as the HMGNN backbone. The dropout rate for the GIN backbone is set to 0.5. The batch size is set as 32 and the number of workers for dataloader is 8. The hidden representations of all levels are 512-dimension.
In the fine-tuning phrase, we implement a 2-layer MLP with ELU activation function as the classifier. The weights of both pre-trained HMGNN and the classifier are optimized by the Adam optimizer with respective learning rates ranging from 1e-4 to 1e-3. For each downstream task, we fine-tune for 100 epochs and report the average test results of five runs. The batch size is 32 and the number of workers is 4. The embedding dimension is 512, the same as that in the pre-training phase. The dropout rate is adjusted in the range [0.5, 0.7]. The parameter settings for each dataset during the fine-tuning process are reported in Supplementary Table 4.
For the purpose of fairness, we pre-train all the baselines on the same dataset ZINC15 according to their official codes, except MoCL28. MoCL does not implement transfer learning, and it is computationally expensive to calculate the similarity matrix of ZINC15 dataset during pre-training. Therefore, we adopt the same manner as the official work: pre-training on each downstream dataset and then fine-tuning on the same dataset for molecular property prediction tasks. All baselines maintain the same split proportion and manner on the downstream datasets.
Data availability
All related data in this paper are public. The ZINC dataset for pre-training can be downloaded from https://github.com/zaixizhang/MGSSL/tree/main/motif_based_pretrain/data/zinc as described in MGSSL30. All downstream datasets for fine-tuning can be downloaded from MoleculeNet website https://github.com/deepchem/deepchem/tree/master/deepchem/molnet/load_function.
Code availability
The implementation of HiMol is publicly available at https://github.com/ZangXuan/HiMol.
References
Kotsias, P.-C. et al. Direct steering of de novo molecular generation with descriptor conditional recurrent neural networks. Nat. Mach. Intell. 2, 254–265 (2020).
Vamathevan, J. et al. Applications of machine learning in drug discovery and development. Nat. Rev. Drug Discov. 18, 463–477 (2019).
LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521, 436–444 (2015).
Li, S., Zhou, J., Xu, T., Dou, D. & Xiong, H. GeomGCL: Geometric graph contrastive learning for molecular property prediction. In Proc. AAAI Conference on Artificial Intelligence, Vol. 36, 4541–4549 (2022).
Wang, Y., Wang, J., Cao, Z. & Barati Farimani, A. Molecular contrastive learning of representations via graph neural networks. Nat. Mach. Intell. 4, 279–287 (2022).
Gebauer, N. W., Gastegger, M., Hessmann, S. S., Müller, K.-R. & Schütt, K. T. Inverse design of 3D molecular structures with conditional generative neural networks. Nat. Commun. 13, 1–11 (2022).
Li, C. et al. Geometry-based molecular generation with deep constrained variational autoencoder. IEEE Trans. Neural Netw. Learn Syst. 1–10 (2022).
Tong, X. et al. Generative models for de novo drug design. J. Med. Chem. 64, 14011–14027 (2021).
Chen, Z., Min, M. R., Parthasarathy, S. & Ning, X. A deep generative model for molecule optimization via one fragment modification. Nat. Mach. Intell. 3, 1040–1049 (2021).
Sutskever, I., Vinyals, O. & Le, Q. V. Sequence to sequence learning with neural networks. In Proc. 27th International Conference on Neural Information Processing Systems, Vol. 2, 3104–3112 (2014).
Kenton, J. D. M.-W. C. & Toutanova, L. K. BERT: Pre-training of deep bidirectional transformers for language understanding. In Proc. 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Vol. 1, 4171–4186 (2019).
Vaswani, A. et al. Attention is all you need. In Proc. 31st International Conference on Neural Information Processing Systems, 6000–6010 (2017).
Weininger, D. Smiles, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 28, 31–36 (1988).
Krenn, M., Häse, F., Nigam, A., Friederich, P. & Aspuru-Guzik, A. Self-referencing embedded strings (selfies): a 100% robust molecular string representation. Mach. Learn. Sci. Technol. 1, 045024 (2020).
Cho, K. et al. Learning phrase representations using RNN encoder-decoder for statistical machine translation. In Proc. 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP) (2014).
Hochreiter, S. & Schmidhuber, J. Long short-term memory. Neural Comput. 9, 1735–1780 (1997).
Zheng, S., Yan, X., Yang, Y. & Xu, J. Identifying structure–property relationships through smiles syntax analysis with self-attention mechanism. J. Chem. Inf. Model. 59, 914–923 (2019).
Xu, Z., Wang, S., Zhu, F. & Huang, J. Seq2seq fingerprint: an unsupervised deep molecular embedding for drug discovery. In Proc. 8th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics, 285–294 (2017).
Xue, D. et al. X-mol: large-scale pre-training for molecular understanding and diverse molecular analysis. Sci. Bull. 67, 899–902 (2022).
Chithrananda, S., Grand, G. & Ramsundar, B. ChemBERTa: large-scale self-supervised pretraining for molecular property prediction. Preprint at arXiv https://arxiv.org/abs/2010.09885 (2020).
Gilmer, J., Schoenholz, S. S., Riley, P. F., Vinyals, O. & Dahl, G. E. Neural message passing for quantum chemistry. In Proc. 34th International Conference on Machine Learning, Vol. 70, 1263–1272 (PMLR, 2017).
Ishida, S., Miyazaki, T., Sugaya, Y. & Omachi, S. Graph neural networks with multiple feature extraction paths for chemical property estimation. Molecules 26, 3125 (2021).
Velickovic, P. et al. Deep graph infomax. ICLR (Poster) 2, 4 (2019).
Qiu, J. et al. Gcc: Graph contrastive coding for graph neural network pre-training. In Proc. 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, 1150–1160 (2020).
You, Y. et al. Graph contrastive learning with augmentations. Adv. Neural Inf. Process. Syst. 33, 5812–5823 (2020).
Xu, M., Wang, H., Ni, B., Guo, H. & Tang, J. Self-supervised graph-level representation learning with local and global structure. In Proc. International Conference on Machine Learning, 11548–11558 (2021).
Liu, S. et al. Pre-training molecular graph representation with 3D geometry. In Proc. 10th International Conference on Learning Representations (2022).
Sun, M., Xing, J., Wang, H., Chen, B. & Zhou, J. MoCL: data-driven molecular fingerprint via knowledge-aware contrastive learning from molecular graph. In Proc. 27th ACM SIGKDD Conference on Knowledge Discovery & Data Mining, 3585–3594 (2021).
Rong, Y. et al. Self-supervised graph transformer on large-scale molecular data. Adv. Neural Inf. Process. Syst. 33, 12559–12571 (2020).
Zhang, Z., Liu, Q., Wang, H., Lu, C. & Lee, C.-K. Motif-based graph self-supervised learning for molecular property prediction. Adv. Neural Inf. Process. Syst. 34, 15870–15882 (2021).
Wang, H. et al. Chemical-reaction-aware molecule representation learning. In Proc. International Conference on Learning Representations (2021).
Zhang, S., Hu, Z., Subramonian, A. & Sun, Y. Motif-driven contrastive learning of graph representations. Preprint at arXiv https://arxiv.org/abs/2012.12533 (2020).
Wang, Y., Magar, R., Liang, C. & Barati Farimani, A. Improving molecular contrastive learning via faulty negative mitigation and decomposed fragment contrast. J. Chem. Inf. Model 62, 2713–2725 (2022).
Zeng, J. & Xie, P. Contrastive self-supervised learning for graph classification. In Proc. AAAI Conference on Artificial Intelligence, Vol. 35, 10824–10832 (2021).
Klicpera, J., Weißenberger, S. & Günnemann, S. Diffusion improves graph learning. In Proc. 33rd International Conference on Neural Information Processing Systems, 13366–13378 (2019).
Velickovic, P. et al. Graph attention networks. In Proc. 6th International Conference on Learning Representations (ICLR, 2018).
Kipf, T. N. & Welling, M. Semi-supervised classification with graph convolutional networks. In Proc. 5th International Conference on Learning Representations (ICLR, 2017).
Xu, K., Hu, W., Leskovec, J. & Jegelka, S. How powerful are graph neural networks? In Proc. International Conference on Learning Representations (2018).
Hamilton, W., Ying, Z. & Leskovec, J. Inductive representation learning on large graphs. Adv. Neural Inf. Process. Syst. 30 (2017).
Liu, M., Gao, H. & Ji, S. Towards deeper graph neural networks. In Proc. 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, 338–348 (2020).
RDkit: Open-source cheminformatics software. https://www.rdkit.org.
Degen, J., Wegscheid-Gerlach, C., Zaliani, A. & Rarey, M. On the art of compiling and using ‘drug-like’chemical fragment spaces. ChemMedChem 3, 1503–1507 (2008).
van der Maaten, L. & Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Liu, Y. et al. RoBERTa: a robustly optimized BERT pretraining approach. Preprint at arXiv https://arxiv.org/abs/1907.11692 (2019).
Ying, C. et al. Do transformers really perform badly for graph representation? Adv. Neural Inf. Process. Syst. 34, 28877–28888 (2021).
Sterling, T. & Irwin, J. J. Zinc 15—ligand discovery for everyone. J. Chem. Inf. Model. 55, 2324–2337 (2015).
Wu, Z. et al. Moleculenet: a benchmark for molecular machine learning. Chem. Sci. 9, 513–530 (2018).
Hu, Z., Dong, Y., Wang, K., Chang, K.-W. & Sun, Y. GPT-GNN: Generative pre-training of graph neural networks. In Proc. 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, 1857–1867 (2020).
Hu, W. et al. Strategies for pre-training graph neural networks. In Proc. International Conference on Learning Representations (2019).
Sun, F.-Y., Hoffman, J., Verma, V. & Tang, J. InfoGraph: Unsupervised and semi-supervised graph-level representation learning via mutual information maximization. In Proc. International Conference on Learning Representations (2019).
You, Y., Chen, T., Shen, Y. & Wang, Z. Graph contrastive learning automated. In Proc. International Conference on Machine Learning, Vol. 139, 12121–12132 (2021).
Acknowledgements
This study is partially supported by the National Natural Science Foundations of China (62276082 and 61876052), Major Key Project of PCL (PCL2021A06), Strategic Emerging Industry Development Special Fund of Shenzhen (20200821174109001) and Pilot Project in 5G + Health Application of Ministry of Industry and Information Technology & National Health Commission (5G + Luohu Hospital Group: an Attempt to New Health Management Styles of Residents).
Author information
Authors and Affiliations
Contributions
Xu.Z. proposed the research, conducted experiments, analyzed the data, and wrote the manuscript. Xi.Z. improved the manuscripts. B.T. supervised the overall project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Chemistry thanks Shengli Jiang and the other anonymous reviewer for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zang, X., Zhao, X. & Tang, B. Hierarchical Molecular Graph Self-Supervised Learning for property prediction. Commun Chem 6, 34 (2023). https://doi.org/10.1038/s42004-023-00825-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42004-023-00825-5
- Springer Nature Limited