Abstract
The alarming increase in global rates of metabolic diseases (MetDs) and their association with cancer risk renders them a considerable burden on our society. The interplay of environmental and genetic factors in causing MetDs may be reflected in DNA methylation patterns, particularly at non-canonical (non-B) DNA structures, such as G-quadruplexes (G4s) or R-loops. To gain insight into the mechanisms of MetD progression, we focused on DNA methylation and functional analyses on intragenic regions of two MetD risk genes, the glucokinase (GCK) exon 7 and the transmembrane 6 superfamily 2 (TM6SF2) intron 2-exon 3 boundary, which harbor non-B DNA motifs for G4s and R-loops.
Pyrosequencing of 148 blood samples from a nested cohort study revealed significant differential methylation in GCK and TM6SF2 in MetD patients versus healthy controls. Furthermore, these regions harbor hypervariable and differentially methylated CpGs also in hepatocellular carcinoma versus normal tissue samples from The Cancer Genome Atlas (TCGA). Permanganate/S1 nuclease footprinting with direct adapter ligation (PDAL-Seq), native polyacrylamide DNA gel electrophoresis and circular dichroism (CD) spectroscopy revealed the formation of G4 structures in these regions and demonstrated that their topology and stability is affected by DNA methylation. Detailed analyses including histone marks, chromatin conformation capture data, and luciferase reporter assays, highlighted the cell-type specific regulatory function of the target regions. Based on our analyses, we hypothesize that changes in DNA methylation lead to topological changes, especially in GCK exon 7, and cause the activation of alternative regulatory elements or potentially play a role in alternative splicing.
Our analyses provide a new view on the mechanisms underlying the progression of MetDs and their link to hepatocellular carcinomas, unveiling non-B DNA structures as important key players already in early disease stages.
Similar content being viewed by others
Introduction
Overweight and obesity represent a major health issue of the twenty-first century. Their worldwide rate tripled from 1975 to 20161, affecting approximately 39% of the global population2. Alarmingly, childhood obesity also increased from 0.7–0.9% in 1975 to 5.6–7.8% in 20163. Caloric imbalance causing obesity is related to dietary habits, physical inactivity, stress, sleep patterns, social and educational status, and endocrine disorders4. Excessive fat accumulation increases the risk of developing metabolic diseases (MetDs) such as the metabolic syndrome (MetS), type 2 diabetes (T2D), metabolic dysfunction-associated fatty liver disease (MAFLD)5,6, and cardiovascular disease (CVD)7. Furthermore, MetD patients have an elevated risk for gastric8, pancreatic9, hepatocellular10, colorectal11, breast12 and lung cancer13,14.
MAFLD is the most common form of chronic liver disease in industrialized societies15 with a worldwide prevalence of 25%16, and is strongly associated with T2D17. One hallmark of MAFLD is the development of large cytoplasmic lipid droplets in hepatocytes with the accumulation of proteins involved in lipid homeostasis on their surface. The key disease mechanism in MAFLD is very likely related to peripheral insulin resistance17,18. In addition, fatty acid β-oxidation is impaired in MAFLD owing to mitochondrial respiratory chain dysfunction, which, in combination with the accumulation of incompletely metabolized fatty acids, contributes to oxidative and lipotoxic stress in hepatocytes. This facilitates the progression from simple steatosis to advanced stages of MAFLD which can further progress to liver fibrosis, cirrhosis, and to hepatocellular carcinoma (HCC)19. Currently, we do not fully understand the molecular mechanisms leading to the development and progression of MAFLD, but they clearly involve a complex interplay between genetic predisposition and environmental factors influencing epigenetic processes, such as DNA methylation at cytosines (5-methylcytosine, 5mC). Studies investigating 5mC have identified differentially methylated positions (DMPs) associated with hepatic fat or key signaling pathways regulating glycolysis and de novo lipogenesis20,21,22,23,24. However, a mechanistic explanation for the impact of those detected DMPs on cellular processes is often lacking. Particularly, if these DMPs are located in intergenic regions or in gene bodies without overlapping regulatory elements, interpreting their biological functions remains difficult.
New insights arise from the observation that G-rich sequences, with the ability to form non-canonical (non-B) DNA structures called G-quadruplexes (G4s), are sites of methylation instability25, and actively influence DNA methylation26. G4s are formed via the consensus motif G≥3NxG≥3NxG≥3NxG≥3 and can adopt intra- or intermolecular planar G4 tetrads held together by Hoogsteen hydrogen bonds with a 1–7 bp long linker of any base (N) and stabilized by a monovalent cation, in vivo typically potassium27,28,29. These structures are found in regulatory regions such as promoters30,31, enhancers32, or near splice sites33, whereas they are less often detected in exons and on the template strand34. They often co-occur with RNA:DNA hybrids (R-loops), formed during the transcription of long non-coding (lncRNAs) on the displaced strand and each stabilizing the other35. G4s act as binding hubs for several transcription factors (TFs)36,37 and DNA methyltransferases (DNMTs)26,38,39, which is underlined by their manifold biological roles extensively reviewed elsewhere40,41,42.
G4s are fairly sensitive to changes in ion concentrations like potassium43, the pH44, or glucose concentrations45. Moreover, DNA methylation influences the thermodynamic properties of the DNA duplex46 and therefore, methylation of CpGs within a G4-forming sequence (G4-CpGs) possibly triggers its rearrangement from a DNA duplex to a non-B DNA structure. This is supported by the observation of enhanced G4 formation due to DNA methylation within the first exon of the human telomere reverse transcriptase (hTERT) gene, which in turn, leads to repressed gene transcription47. Furthermore, DNA methylation leads to increased stability of G4 structures in BCL248, VEGF49 and FMR150, whereas decreased stability was detected for the MEST G451 in in vitro assays. Even changes in the G4 topology from parallel to antiparallel in c-KIT, or a complete loss of G4 formation in HRAS can be caused by DNA methylation52. Genome-wide approaches revealed that methylation of G4-CpGs is significantly decreased compared to CpGs outside of G4s, especially among those CpGs located in CpG islands (CGIs)26,53. Furthermore, it has been shown, that DNMT1 preferentially binds to G4 structures rather than to hemi-methylated B-DNA, which results in the inhibition of the DNMT1 and local hypomethylation at the G4 structure26. There are still contradictory discussions, since the mentioned studies mostly have shown that G4-CpGs are targets of hypomethylation, while a recent work investigating DNA methylation of aging clocks suggests a general DNA methylation instability at G4-CpGs, rather than a particular hypo- or hypermethylation25. Furthermore, whereas studies investigating individual G4 motifs have shown that DNA methylation can also stabilize G4s, genome-wide analysis demonstrated that the stability of G4s with a maximum distance of 50 nucleotides to the CGI is associated with hypomethylation and in cases where the G4-containing CGI is located in closed and hypermethylated chromatin, the G4 stability was lowest54. Most analyses focused on promoters and on regions overlapping CGIs, but a considerable number of regulatory regions, like alternative promoters55,56 or enhancers57, occur outside of CGIs where different regulatory mechanisms probably apply. Selective DNA methylation loss in CG-poor regions can make chromatin accessible for alternative transcription, particularly in HCC58, and we have previously associated differential methylation with alterations in promoter activation59. This indicates that we are still in the early stages of understanding the mechanisms involved, and that different genetic locations probably underlie distinct mechanisms. Therefore, changes in the G4 landscape or their topology could alter transcription factor binding and activate alternative regulatory elements or alternative splicing events, causing aberrant gene expression patterns that contribute to MAFLD and may even promote the development of more severe diseases such as hepatocellular carcinoma.
Patients suffering from metabolic diseases are at a higher risk of developing cancer during their lifetime. Therefore, it is necessary to understand which processes are involved and DNA methylation at G4s could be one of these mechanisms, probably already arising in a non-cancerous but dysregulated metabolic stage. Hence, we aimed to analyze DNA methylation and G4 formation in MetD patients and in HCC in two intragenic regions outside of CGIs with annotated regulatory elements, as well as the occurrence of putative G4 forming motifs and lncRNAs in the key metabolism genes glucokinase (GCK) and transmembrane 6 superfamily 2 (TM6SF2). GCK deficiency has been linked with T2D60 and neonatal diabetes61, while a common GCK SNP has been associated with hyperglycemia62 and several alternative GCK promoters are annotated for the gene body. Differential methylation of the GCK CGI spanning exons 9–10 had been linked with CVD63. The intron 2-exon 3 boundary in TM6SF2 overlaps with a prognostic lncRNA for cancer64,65, it shows many different transcripts and rs58542926 C > T, the SNP prognostic for MAFLD66 and HCC67, is located downstream of the annotated regulatory region. rs58542926 C > T is routinely checked in the clinics and potentially could have an impact on the methylation of the next regulatory element.
Herein, we report differential DNA methylation in exon 7 of GCK and within the intron 2-exon 3 boundary of TM6SF2 in blood samples from a MetD patient cohort in G4 forming regions, which are involved in the activation of alternative regulatory elements and possibly also in alternative splicing. These new insights emphasize the critical need to understand the interplay between aberrant DNA methylation and non-B DNA structure formation occurring already at an early disease stage.
Results
Cohort demographics and description
To investigate DNA methylation in putative intragenic G4-forming regions in GCK and TM6SF2, we performed locus-specific 5mC analysis on the Qiagen Pyromark Q24 Platform for 148 blood donors which were classified as lean-healthy (LH), obese-healthy (OH), lean-MAFLD (LM), obese-MAFLD (OM), and advanced MetD (aMetD, including MAFLD and poorly controlled T2D patients) according to their metabolic parameters, as described in the Methods section. The mean values of the clinical parameters are provided in Table 1 and Supplement Fig. 1. The age of our study cohort ranged from 42–77 years, albeit the OM and aMetD groups showed significantly elevated age compared to the LH group (LH vs. OM p = 0.0152, LH vs aMetD p = 1.53e-05, Student´s t-test, and Supplement Table 1). This trend was not unexpected since MAFLD and T2D are known as aging-related diseases occurring at a higher frequency in the elderly (reviewed in68). Fasting blood glucose, insulin, and aspartate transferase (AST) levels were significantly increased in OH, OM and aMetD, but not in LM compared to LH (Students t-test, Supplement Table 1). No significant change was detected in total cholesterol levels in any group compared to healthy controls. High-density lipoprotein (HDL) was significantly decreased in OM and aMetD, and low-density lipoprotein (LDL) was only decreased in aMetD, whereas total triglycerides were increased in LM, OM, and aMetD. The detailed p-values are shown in the Supplement Table 1.
BMI Body mass index, HOMA homeostasis model assessment, HbA1c Glycated haemoglobin A1c, FIB4 Fibrosis4 score, Fast. Glucose Fasting glucose, Fast. Insulin Fasting insulin, GGT γ-glutamyl transpeptidase, AST aspartate aminotransferase, ALT alanine aminotransferase, HDL-c High density lipoprotein cholesterol, LDL-c Low density lipoprotein cholesterol, TG Triglycerides, TChol total cholesterol;
GCK exon 7 and the TM6SF2 intron 2-exon 3 boundary are differentially methylated in MetD patients and are hypervariable in HCC
GCK is encoded on the minus strand of chr. 7 and consists of ten exons (Fig. 1A). 14 different transcripts are annotated for GCK in the ENSEMBL genome browser69. The investigated region is located in exon 7 (chr7:44,187,339–44,187,380; hg19) and is overlapped by an antisense lncRNA LOC105375258 (XR_927223.3) on the plus strand. Genome segmentation by automating chromatin state discovery and characterization (ChromHMM) in the UCSC genome browser predicts a “weak enhancer or open chromatin cis regulatory element” in hepatoblastoma cell line HepG2 and a “promoter region including TSS” in the lymphoblast cell line K562 (Fig. 1A, yellow rectangle)70. Furthermore, several alternative promoters are found in intron 7 and 8, as well as an EGR1 TF binding site (Fig. 1A, green rectangle)70. Significant hypomethylation was detected in CpG 1 and 2 in aMetD patients, and in CpG 3 in LM, OM and aMetD compared to LH subjects. No changes were detected in CpG 4 (Fig. 1A lower panel and Supplement Fig. 2A). In particular, a gradual reduction in DNA methylation was detected for CpG 3 with the individual disease stages, and the advanced MetD patients showed the lowest methylation.
DNA methylation in GCK exon 7 and within the TM6SF2 intron 2-exon 3 boundary. Detailed information about the differentially methylated regions in (A). GCK and (B). TM6SF2 including the nearest annotated CpG positions on the Illumina Methylation array. The yellow rectangle indicates the genome segmentation by ChromHMM in UCSC genome browser70. The green rectangle represents the EGR1 binding motif. The pyrosequencing region is marked in red. The lower panel shows DNA methylation at the single CpG resolution by metabolic status in A. for GCK CpG 1–4 and B. TM6SF2 CpG 3–6. DNA methylation data of the nearest annotated CpG on the Illumina Methylation array for (C). GCK cg23381646 and (D). TM6SF2 cg11911689 are shown comparing HCC to normal tissue from TCGA71. Gene expression for (E). GCK, (F). TM6SF2 and (G). the TM6SF2-overlapping lncRNA ENSG00000267629 were obtained from the liver expression atlas (GepLiver)72. Pairwise comparison using Students t-test against LH (A.-B.) and Wilcoxon test (C.-G.), *p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
In line with GCK exon 7, we found differential methylation also for the TM6SF2 intron 2-exon 3 boundary (chr19:19,381,224–19,381,297; hg19, Fig. 1B). TM6SF2 is encoded by ten exons on the minus strand of chr. 19. In total, there are five transcripts annotated, of which two result in a retained intron and one in a skipped exon overlapping our target region69. Notably, exon 3–5 are targets of alternative splicing events and exon 3 is annotated as cassette exon (Supplement Fig. 3)70. Furthermore, TM6SF2 overlaps with the sense lncRNA ENSG00000267629 (AC138430.4), sharing the same transcription start site. This lncRNA represents a readthrough between TM6SF2 and HAPLN4. ChromHMM predicts a “weak enhancer or open chromatin cis regulatory element” in HepG2 and a “promoter region including TSS” in K562 in the UCSC genome browser70, as well as an EGR1 binding site (Fig. 1B). 5mC analyses revealed that CpG 3–5 were significantly hypermethylated in the advanced MetD group. In contrast, CpG 4 and 5 were significantly hypomethylated in OM compared to LH and CpG 5 was also significantly hypomethylated in LM compared to LH (Student´s t-test, Fig. 1B lower panel and Supplement Fig. 2B).
Given that alterations in DNA methylation are also associated with aging73 and metabolic diseases occur at a higher frequency with increasing age, we analyzed the magnitude of age-relation compared to published aging markers located in EDARADD (cg09809672), IPO8 (cg19722847), NHLRC1 (cg22736354), P2RXL1 (cg05442902) and SCGN (cg06493994). These five markers were selected based on their identification in three distinct epigenetic clock studies utilizing Illumina Methylation arrays73,74,75. Consistent with published data, we detected a significant positive (NHLRC1 and SCGN) and negative (IPO8, EDARADD and P2RXL1) association between DNA methylation and chronological age in our cohort (Supplement Fig. 4). In addition, the correlation analysis showed a weak but significant age-dependency for GCK CpG 3 and TM6SF2 CpG 1, 3 and strongest for CpG 5, as well as for the mean DNA methylation (mean of CpG 1–6). Therefore, we restricted our analysis to 42–60-year-old subjects. We still observed a significant decreased DNA methylation at GCK CpG 3 in all groups compared to lean-healthy subjects, which reached significance even in obese-healthy patients, while this was not the case when analyzing the entire age-spectrum (Supplement Fig. 2C). TM6SF2 CpG 3, 4 and 6 where still significantly differentially methylated, but the effect was lost in CpG 5 in 42–60-year-old subjects (Supplement Fig. 2D). This points out that aberrant DNA methylation in these regions is only partly caused by aging processes and underlies also a disease-related mechanism.
Next, we were curious if the SNP rs58542926 C > T, which causes a p.Glu167Lys (E167K) substitution in TM6SF2, could influence DNA methylation in this specific regulatory region. The SNP is located 1,696 bp away from CpG 4 in exon 6. Although the risk associated variant T is more common in the advanced MetD group (Supplement Fig. 5A), we found no significant difference in DNA methylation by genotype in our study cohort (Supplement Fig. 5B). Therefore, we can exclude that the genetic variant rs58542926 C > T causes the detected differential DNA methylation within TM6SF2 intron 2-exon 3.
Since metabolic diseases are associated with an increased risk for HCC10, we investigated 5mC of GCK and TM6SF2 in data of The Cancer Genome Atlas (TCGA) from HCC compared to normal tissue71. The nearest annotated CpG for GCK covered by the Illumina Methylation array is cg23381646 located 45 bp upstream of CpG 3 (Fig. 1A). Although DNA methylation at cg23381646 was not significantly different (Wilcoxon test, Benjamini-Hochberg (BH) adjusted p = 0.58, Fig. 1C), it appeared as highly variable in tumor tissue with DNA methylation beta-values ranging from approximately 0.25–0.8.
For TM6SF2, TCGA data showed that the nearest annotated CpG cg11911689 (78 bp downstream of CpG 4, Fig. 1B) was significantly hypomethylated in HCC compared to normal tissue (Wilcoxon test, BH adjusted p = 4.5E-10, Fig. 1D), and again showed a highly variable DNA methylation range in cancer compared to normal control tissue.
To explore gene expression of the target genes, we analyzed GCK and TM6SF2 expression in MAFLD and HCC in the liver expression database (GepLiver)72, since transcriptome data were not available for the MetD cohort due to the lack of liver material. GCK expression was increased by 1.73-fold (Wilcoxon test, BH adjusted p = 2.52E-32) in MAFLD, but decreased expression was detected in HCC compared to normal liver tissue (0.67-fold, Wilcoxon test, BH adjusted p = 2.73E-32, Fig. 1E). Significant increased expression was also detected for TM6SF2 in MAFLD (1.04-fold, Wilcoxon test, BH adjusted p = 0.035), which was again decreased in HCC by 0.76-fold (Wilcoxon test, BH adjusted p = 7.48E-28, Fig. 1F). Of note, the expression of the TM6SF2 overlapping lncRNA ENSG00000267629 was increased in MAFLD by 1.92-fold (Wilcoxon test, BH adjusted p = 4.31E-7). Although it remained slightly overexpressed in HCC, it did not reach significance (1.08-fold, Wilcoxon test, BH adjusted p = 0.58, Fig. 1G).
Differentially methylated regions detected in GCK and TM6SF2 are able to form non-B DNA structures
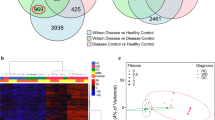
To investigate if the detected DMRs are located in non-B DNA structures and if they are formed in living cells, we retrieved information about R-loop forming sequences (RLFS)76 and putative G4 motifs (PQS) from pqsfinder77. The GCK DMR harbors several G4 motifs, but no predicted RLFS, albeit this exon is overlapped by the annotated antisense lncRNA LOC105375258 (Fig. 2A left panel). All four analyzed CpG positions in GCK exon 7 are located in a G4 motif in the non-template strand, with the top differentially methylated CpG 3 located in the longest loop of the G4 between the G-run two and three. Notably, the G4 includes a bulge, represented as an interruption in the G-run, between the third and fourth G of G-run two (Sequence motif in Fig. 2A left panel). Furthermore, the downstream exons 8–10 harbor both RLFS and PQS (Fig. 2A left panel). DNA methylation of this downstream region was investigated elsewhere and showed also differential methylation in cardiovascular diseases63. The TM6SF2 DMR contained a RLFS as well as PQS (Fig. 2A middle panel). The differentially methylated CpGs 4–6 were located directly within the predicted G4 motif in the template strand and CpG 1–3 flanked the G4 motif. As negative control region we have chosen an intronic region of PARD3B without annotated PQS or RLFS (Fig. 2A right panel). In vitro G4 formation was verified by analyzing published G4-sequencing (G4-Seq) maps based on a polymerase stop assay in a buffer containing the G4 stabilizing ligand pyridostatin (PDS) in HEK293T cells (Fig. 2A, black track)78. Strong signal intensities in GCK and TM6SF2, but not in PARD3B, confirmed the ability of the predicted motifs to form DNA secondary structures. As G4s are not necessarily formed in living cells and their formation varies strongly in different cell types, we analyzed published G4 ChIP-Seq data in HEK293 cells79. Especially, GCK shows enriched signal intensities compared to its input control over the whole region spanning from exon 7 to exon 10, rather than distinct peaks similar to the G4-Seq signals (Fig. 2A left panel). TM6SF2 showed a more discrete peak for replicate 1 but not for replicate 2 (Fig. 2A middle panel). Since, we could not infer a clear picture for G4 formation from the antibody-based ChIP-Seq data, particularly for GCK exon 7, we aimed to establish an alternative chemical method based on permanganate/S1 nuclease footprinting80 which in general detects regions of single-stranded DNA (ssDNA). We updated this method by linking it to direct adapter ligation on streptavidin-coated beads, followed by an on-bead PCR published as PDAL-Seq (permanganate/S1 nuclease footprinting with direct adapter ligation and sequencing)81. Since PDAL-Seq is a footprinting method, we expected low or absent read frequencies directly at the non-B DNA structure flanked by increased read frequencies at the non-B DNA borders. For HEK293 cells, we did not detect a direct overlap of PDAL-footprints with G4-Seq peaks at GCK exon 7 rather than a slight upstream shift, but in MCF7 cells the non-B-footprint was more obvious, highlighting the cell type differences (Fig. 2A left panel, light blue highlighted box). Of note, footprints detected in GCK exon 8–10 were more pronounced and overlapped well with the strong G4-Seq signals, while the peaks from the antibody-based G4 ChIP-Seq method were less clear (Fig. 2A, left panel, light red highlighted box). In contrast, the intronic region of PARD3B did not contain a G4 motif, nor did it form G4s in vitro or in living cells (Fig. 2A right panel).
Non-B DNA formation in GCK and TM6SF2: (A). Shows GCK (chr7: 44,184,901–44,187,919, hg19), TM6SF2 (chr19: 19,380,358–19,382,362, hg19) and the non-G4 forming region in PARD3B (chr2:205,780,593–205,782,891, hg19). Data from different sources are shown in individual rows: Pink rectangles indicate R-loop forming sequences (RLFS) calculated by Quantitative Model of R-loop Forming Sequence (RLFS) finder (QmRLFS-finder)76. Blue rectangles represent putative G4 forming motifs (PQS) obtained from pqsfinder77, Black track: in vitro G4 formation stabilized with pyridostatin (PDS) in HEK293T cells78; Grey track: ChIP input control79; Red tracks: G4-ChIP-Seq tracks in HEK293 cells (2 replicates)79; Blue tracks: PDAL-Seq replicates 1 and 2 of HEK293 and MCF7 cells, Olive tracks: transcription factor EGR1 binding in K546 cells82. The sequences within GCK (antisense G4, sequence is given as reverse complement) and TM6SF2 (sense G4) forming a G4 structure are detailed below. G-tracks are underlined, grey boxes indicate CpG positions. Blue highlighted areas: identified differentially methylated regions; light red highlighted area: a second regulatory region in GCK, which can form G4s. (B). Native DNA PAA gels identify in vitro G4 formation in the G4 control region in MYC, GCK exon 7 and in the TM6SF2 intron 2-exon 3 boundary, but not for ESR1 (negative control). PCR products are loaded as unfolded (50 mM Tris–HCl pH 8.3) (U), folded with 150 mM KCl and heat induction (K), and folded with 150 mM KCl, heat induction and 10 µM PDS (PDS). (C). Positive and negative peak maxima obtained from CD spectra for MYC, GCK and TM6SF2 with increasing K + concentrations, for the methylated or mutated oligos. Individual CD spectra are shown for (D). MYC, (E). TM6SF2 and (F). GCK. (G). Depicts the possible refolding of the methylated hairpin to a parallel G4 structure caused by demethylation at the GCK exon 7 region. The green loop indicates the bulge sequence which interrupts G-run two.
Next, we analyzed EGR1 ChIP-Seq data in K562 from ENCODE82. Unfortunately, there were no data available in HEK293 or MCF7 cells. Strong EGR1 signals were detected co-occurring with the G4-Seq peak and the footprinting valley of PDAL-Seq. Currently we do not know, if non-B DNAs are formed in K562 cells. The negative G4 region in PARD3B showed also a EGR1 binding site indicating that EGR1 does not only target G4 regions.
To independently evaluate the G4-forming ability of the target regions, G4 folding was confirmed by native DNA polyacrylamide (PAA) gel electrophoresis (Fig. 2B). The GCK and TM6SF2 target regions and two controls, MYC as a positive G4-folding region and an enhancer region of ESR1 as a G4-negative region, were PCR amplified and separated on a native 10% DNA PAA gel. Strikingly, both GCK and TM6SF2, as well as MYC showed significant shifted bands after the G4 folding procedure with potassium and heat induction (Fig. 2B, labeled with K), and even stronger after G4-stabilization with PDS (Fig. 2B, labeled with PDS), compared to their unfolded controls only containing Tris-buffer (Fig. 2B, labeled with U). This effect was not detected for the negative control region ESR1. In agreement with the native gel electrophoresis, circular dichroism (CD) spectroscopy experiments (Fig. 2C–F) showed G4 formation for all regions enhanced by increasing K+ concentrations. MYC (Fig. 2C,D) and TM6SF2 (Fig. 2C,E) showed characteristic parallel G4 structures with a negative peak at 240 nm and a positive peak at 260 nm. DNA methylation within MYC did not change the conformation of the G4 nor did it lead to a loss of the formed structure. For GCK we noticed an interesting pattern. While the increased K+ concentrations led to parallel G4 formation visible due to a shift in the positive peak from 279 to 267 nm, the methylated GCK oligo showed a broad positive peak from 263-285 nm, which resembled a hairpin structure (Fig. 2D,F). This conformational change was probably caused by the methyl-groups which destabilize the G4, since there were six bases within the bulge followed by one G belonging to G-run two and 16 bases in loop two, which are able to pair (Fig. 2G). In all CD-spectra, the mutated oligos showed a clear shift of the positive peak to around 280 nm, indicating the loss of G4 formation.
Differentially methylated regions in GCK and TM6SF2 harbor regulatory functions and contact neighboring genes
To assess the regulatory potential of the identified DMRs located in G4-forming regions in GCK and TM6SF2, we analyzed the activating H3K27ac, H3K4me1, H3K4me3 and the repressing H3K27me3 histone marks in H1-hESC (human embryonic stem cell line, red), HepG2 (hepatoblastoma cell line, blue), NHLF (lung fibroblasts, orange), K562 (lymphoblasts, brown), NHEK (normal human epidermal keratinocytes, violet), and MCF7 (breast cancer, green) cell lines in IGV from the ENCODE project82. In GCK exon 7 H3K4me1 activating histone marks were found in K562 cells and to a lesser extent also in HepG2 cells, pointing towards and enhancer function (Fig. 3A, left panel). NHLF and NHEK cells harbored the repressive H3K27me3 mark and MCF7 cells showed H3K27ac marks near the target region in exon 7. Furthermore, the downstream regions overlapping exon 8–10 showed H3K4me1 in HepG2 and K562 cells, but H3K27me3 in H1-hESC, NHLF and NHEK cells. For the TM6SF2 intron 2-exon 3 boundary (Fig. 3A, right panel) we found strong signals for the activating H3K4me1 histone modification in K562 cells, but H3K27me3 signals in NHEK and NHLF cells. HepG2 cells showed H3K4me1 and H3K4me3 marks in the promoter region of TM6SF2. In conclusion, the analysis of ENCODE histone modifications revealed cell type-specific regulatory elements for both target regions.
Regulatory roles of GCK and TM6SF4 target regions. (A). Activating (H3K27ac, H3K4me1 and H3K4me3) and repressing (H3K27me3) histone modifications obtained from ChIP-seq experiments from ENCODE for H1-hESC (stem cells, red), HepG2 (hepatoblastoma, blue), NHLF (healthy lung, orange), K562 (blood, brown), NHEK (normal human epidermal keratinocytes, violet) and MCF7 (breast cancer, green) cell lines82 in GCK (left panel) and TM6SF2 (right panel). For the hg19 genome assembly ENCODE did not provide H3K4me1 data for MCF7 cells. Yellow highlighted areas represent the pyrosequencing regions. Here we showed a cell-type specific pattern of activating and repressing histone marks. (B). The CpG-free basic vector was used to test for a putative promoter function and the CpG-free promoter (containing a minimal promoter) vector to test for a putative enhancer function of the differentially methylated regions in a luciferase gene reporter assay in (C). GCK exon 7 and (D). in the TM6SF2 intron 2-exon 3 boundary, in a methylated and unmethylated state compared to the empty vector (EV). Pairwise comparison against empty vectors (Welch two sample t-test), *p < 0.05, **p < 0.01, ***p < 0.001.
To analyze the effect of the identified differentially methylated regions on gene expression, we performed in vitro luciferase reporter assays. Therefore, we cloned the regions of interest in two different CpG-free vectors upstream of the luciferase reporter gene and tested for their potential promoter and enhancer functions in a methylated and unmethylated state (Fig. 3B). We detected both, a significant promoter (60.3±11.3-fold, Welch two sample t-test, p = 0.0004,) and enhancer (27.4±3.9-fold, Welch two sample t-test, p = 0.0001, Fig. 3C) function for the GCK region compared to the empty vector, reflecting well the annotated ChromHMM function in HepG2 in the UCSC genome browser. However, when the same region was methylated prior to the analysis, both functions were lost. In contrast, we did not find a promoter function for the DMR located in TM6SF2, but the methylated construct showed a 1.7 ± 0.2-fold (Welch Two Sample t-test p = 0.004, Fig. 3D) elevated enhancer function.
To predict the potential neighboring targets of the identified GCK and TM6SF2 regulatory regions, we analyzed Hi-C data in HepG2 cells82 and from HCC patient material83 using the 3D Genome Browser84. GCK is located in a topological associated domain (TAD) of approximately 500 kb. Strikingly, within this TAD several genes are known for their association with metabolic diseases and cancer like AE binding protein 1 (AEBP1)85,86, YKT6 v-SNARE homolog (YKT6)87,88, upregulator of cell proliferation (URGCP)89,90 and phosphoglycerate mutase 2 (PGAM2; for more details see Supplement Sect. 4)91. When comparing the interaction frequencies of HepG2 cells and HCC decreased signal intensities were observed (Fig. 4A, comparison between TAD interaction frequency in HepG2-top and HCC-bottom).
HiC data analysis in HepG2 cells and in HCC tumor material visualized in 3D genome browser84. (A). TAD formation on chr. 7 around the GCK DMP in HepG2 (upper panel)82 and HCC tumor tissue (lower panel)83. (B). Analysis of potential contact points of the regulatory region (red vertical line) shows interactions (blue highlighted areas) with GCK intron 1 and an intergenic region between MYL7 and POLD2 near an enhancer region. (C). TAD formation on chr. 19 around TM6SF2 in HepG2 (upper panel)82 and HCC tumor tissue (lower panel)83. (D). Analysis of potential contact points of the regulatory region (red vertical line) shows interactions (blue highlighted areas) with SUGP1 and HAPLN4. Chromosome annotations are given in the newer assembly hg38.
By analyzing the potential contact points of the GCK DMP, we noticed that this region stays in contact with an enhancer region in GCK intron 1 (~ chr7:44,157,293–44,158,172; hg38) and a proximal enhancer region between myosin light chain 7 (MYL7) and DNA polymerase delta 2 (POLD2, Fig. 4B). TM6SF2 is located in a TAD together with UPF1 RNA helicase and ATPase (UPF1) and neurocan (NCAN, Fig. 4C), associated with MAFLD92 and HCC93. The interaction analysis showed that the TM6SF2 DMP contacts a promoter-like region within intron 3 of hyaluronan and proteoglycan link protein 4 (HPLN4, ~ chr19:19,260,457, hg38) as well as intron 8 of SURP and G-patch domain containing 1 (SUGP1, ~ chr19:19,280,457, hg38), which was identified as regulator of cholesterol metabolism (Fig. 4D)94.
In summary, both in silico data for histone marks and HiC data, as well as the luciferase reporter assay provide evidence that GCK exon 7 and the TM6SF2 intron 2-exon 3 boundary harbor important regulatory elements, which are not only related to metabolic diseases, but also could play roles in tumor formation and prognosis. Furthermore, our analysis points out that these regions contact neighboring genes and that changes in their methylation patterns potentially also affect the interactions with these elements. This strongly indicates that changes in DNA methylation at these regulatory elements are not only associated with MAFLD but could also be linked to the enhanced cancer risk in MetD patients corroborated also by the changes in the HiC data within these TADs.
Discussion
Metabolic diseases often arise due to a complex interplay of genetic and epigenetic processes. Whereas the genetic risk is predefined by the base composition, the epigenetic landscape is flexible and heavily influenced by the individuals´ environment. To assess epigenetic changes in two metabolic disease associated genes, GCK and TM6SF2, we analyzed 5mC in 148 patients grouped in five different categories: lean- and obese healthy subjects, lean- and obese MAFLD, as well as patients suffering from advanced MetDs. We focused on two intragenic regions, annotated as regulatory regions in ChromHMM and predicted as non-B DNA forming sites (G4s and R-loops), which are not overlapping with CGIs. Significant 5mC changes were detected for both target genes. While GCK exon 7 was significantly hypomethylated especially at CpG 3 in all groups compared to lean healthy subjects (Fig. 1A), we observed a diverse pattern for the TM6SF2 intron 2-exon 3 boundary. Lean- and obese MAFLD subjects showed hypomethylation, whereas hypermethylation was detected in advanced MetD (Fig. 1B). This pattern likely mirrors the elevated blood glucose levels observed also as HbA1c > 7% within the advanced MetD group, potentially leading to hypermethylation. Furthermore, it also could indicate a change in blood cell type composition in this group, which also influences DNA methylation patterns95.
Since increasing chronological age is one major risk factor for developing metabolic diseases (reviewed in68), our findings of a significant age correlation with DNA methylation within GCK and TM6SF2 were expected. Nevertheless, after limiting the analysis to patients between 42–60-years of age, significant differential methylation was still detected for both genes, pointing towards a combined disease and aging mechanism. In addition, we were able to exclude an influence of the TM6SF2 disease risk SNP rs58542926 C > T on DNA methylation in this region.
Moreover, we confirmed our results in hepatocellular carcinoma versus adjacent normal tissue obtained from TCGA data71. Although in contrast to cg11911689 (TM6SF2, Fig. 1D), differential methylation was not significant in cg23381646 (GCK, Fig. 1C), both CpGs showed a highly variable 5mC range, which is a sign for DNA methylation instability as previously observed, e.g. in Alzheimer’s disease96. Furthermore, DMPs in TM6SF2 were hypermethylated in advanced MetD patients, whereas the TCGA data from HCC tissue material showed hypomethylation. The observation of opposing directions of differential methylation in blood versus the affected tissue is not unusual. In our latest epigenome-wide association study, a regulatory region upstream of NHL repeat containing 1 (NHLRC1) was found to be hypervariable, and hypermethylated in blood of lung cancer patients, but hypomethylated in lung cancer versus normal adjacent tissue97. Although blood DNA methylation patterns are not necessarily reflecting the changes in the affected tissues, especially in metabolic diseases where blood glucose and triglyceride or cholesterol levels are constantly elevated when untreated, these patterns could direct us towards the affected target genes.
The analysis of gene expression in liver tissue of MAFLD and HCC patients demonstrated their differential expression. In line with the data from the liver expression database (Fig. 1E–G)72, Ilieva and colleagues found that GCK expression was increased by 2.48-fold (false discovery rate (FDR) adjusted p = 1.08E-05) in MAFLD compared to obese-healthy subjects and by 1.83-fold (FDR adjusted p = 1.21E-05) in advanced MetD patients compared to lean-healthy subjects in blood98. In contrast, GCK was downregulated in HCC compared to normal tissue from TCGA, which is at least in part explained by the replacement of GCK by hexokinase 2 in tumor cells99. The same trend was observed for TM6SF2, which was overexpressed in MAFLD, but decreased in HCC. Strikingly, the overlapping sense lncRNA ENSG00000267629 was overexpressed by 1.92-fold in the liver expression database in MAFLD compared to normal tissue72 and even by 3.19-fold (FDR adjusted p = 0.0005898) in MAFLD compared to obese-healthy subjects in the study of Ilieva et al.98. Furthermore, the expression level of this lncRNA was prognostic for the survival of intrahepatic cholangiocarcinoma (iCCA) with a hazard ratio (HR) of 1.582 (1.143–2.190, 95% CI, p = 0.006). Patients with a higher survival probability showed lower expression of this lncRNA transcript65. Moreover, it was also prognostic for patients suffering from acute myeloid leukemia (AML)64.
Summarizing, DNA methylation of the target regions is not only detected in early stages of a metabolic disease (in TM6SF2 after age correction even already in obesity) but also in HCC, indicating that DNA methylation changes observed in cancer are in some cases already detectable in a precancerous state which is in keeping with the increased risk for HCC observed in MAFLD patients.
G4s have emerged as major contributors to DNA methylation regulation and therefore, the detection of DMPs in G4 forming motifs is not new. Nevertheless, the shared occurrence of aberrant DNA methylation in an early disease stage such as obesity or metabolic diseases prior to cancer formation, implies an urgent need to understand their functions. We were able to show G4 formation in the TM6SF2 intron 2-exon 3 boundary with PDAL-Seq in living cells (Fig. 2A), which adopts a parallel structure according to the CD-spectra (Fig. 2E). Overall, the DNA methylation in normal liver tissue (Fig. 1D) and also in several cell lines is low (Supplement Fig. 6A). Therefore, stable G4 formation is most likely occurring in this region. In contrast, GCK exon 7 harbors a ssDNA stretch, but slightly downstream of the G4-Seq peak. Since many cell lines such as HepG2 or MCF7 (Supplement Fig. 6B) as well as the normal liver tissue (Fig. 1C) are highly methylated in this region, stable G4 formation is unlikely, rather than a hairpin, as inferred from the CD spectra of the methylated GCK oligo. Hence, when demethylation occurs, a parallel structure is formed (Fig. 2F,G). This observation is in line with the structural change in c-KIT and HRAS caused by DNA methylation52. Furthermore, PDAL-Seq revealed the formation of G4 structures also exon 8–10, which has been shown as significantly hypomethylated in patients suffering from coronary heart disease (49.77 ± 6.43%) compared to controls (54.47 ± 7.65%, p = 0.018)63. CVDs are a common comorbidity of MAFLD and T2D and therefore this hypomethylation reflects the same pattern as the observed hypomethylation at exon 7 in our cohort.
When differential methylation occurs within regulatory elements, it inevitably affects gene expression patterns. Both regions exhibit cell-type-specific histone marks (Fig. 3A), luciferase reporter assays demonstrate that their regulatory functions are activated in a DNA methylation-dependent manner (Fig. 3B,C), and HiC data indicate that these elements interact with neighboring genes (Fig. 4). Notably, DNA methylation of both regions is highly variable, and exhibit enriched binding for the zinc finger transcription factor EGR1. While it has been reported that G4s drive DNA methylation processes by sequestering DNMT1 to the G4 and cause its inactivation26, we propose a slightly different, DNMT1-independent mechanism for these regions.
EGR1 recognizes a G-rich motif100 and is predicted as G4 binding protein101. It recruits the ten eleven translocase 1 (TET1), leading to DNA demethylation processes adjacent to its binding sites102,103. Its binding is likely not restricted solely to folded G4s, as the PARD3B region, which lacks any non-B DNA structure, also shows EGR1 binding. EGR1 has been shown as important transcription factor regulating inflammatory genes by binding to their enhancers104. EGR1 is upregulated in MAFLD, and a its knockdown resulted in a reduced expression of pro-inflammatory genes in hepatocytes105.
Given that the GCK exon 7 G4-CpGs are methylated in normal liver tissue, as well as different cell lines, we hypothesize that local demethylation processes via TET1 initiated by EGR1 binding facilitate G4 formation, thereby allowing transcription factor binding and activation of the regulatory element (Fig. 5A). This is supported by experiments showing a ~ 60-fold increase in promoter activity and a ~ 27-fold increase in enhancer function when GCK exon 7 is unmethylated. The TM6SF2 intron 2-exon 3 boundary shows clear non-B DNA formation, and methylation reduces the stability of its G4 structure. DNA methylation at the nearest CpG position is lower in the target region in TM6SF2 than in GCK in both normal tissues and cell lines, implying that this element is active in a healthy state, at least in a cell-type specific manner. The strong binding of EGR1 to the region suggests that DNA demethylation is facilitated by EGR1 and TET1 recruitment and a stable parallel G4 is formed. However, increased DNA methylation in this region, as seen in advanced MetD patients or in cases of high methylation instability as observed in HCC, could impact the regulatory element's activity. Notably, we analyzed only a rather small part of the annotated regulatory element, which may have caused the overall low detected enhancer strength. In line with our study, but investigating long G4-forming regions, Williams et al. found that G4-forming regions harbor promoter and enhancer functions and that the most prominently enriched interacting transcription factor binding site was that of EGR132.
Mechanistic explanation. (A) Hypomethylation of GCK exon 7 leads to the formation of a G4 structure which in turn activates the alternative regulatory element. (B) Methylation instability at the cassette exon and G4 forming region in the TM6SF2 intron 2-exon 3 boundary could cause polymerase slow down or stalling and alternative splicing events leading to a readthrough of TM6SF2 to HAPLN4, forming the long non-coding RNA. Created with BioRender.com.
Alternative promoters play critical roles in hepatocellular and breast cancer, two cancer types highly linked to metabolic diseases and obesity10,12. Genes involved in glycolysis and cholesterol metabolism show elevated expression from alternative promoters, which typically exhibit low H3K4me3 levels and hypomethylation in CpGs located in non-CGI promoters58. CGIs are underrepresented in alternative promoters, and Nepal et al. propose that activation of these promoters in HCC occurs through demethylation of CpG-poor intragenic regions58, similar to the investigated G4-forming regions.
Additionally, the TM6SF2 intron 2-exon 3 boundary could be subject to alternative splicing, since exon 3 of TM6SF2 is annotated as a cassette exon, and G4s are enriched at distal splice sites of such exons33. Splicing factors like U2AF1, which recognizes the 3′ splice site, are among the top candidates associated with G4s37. G4 formation regulates alternative splicing of BCL-x, since stabilization of the G4 structure promotes the expression of the pro-apoptotic BCL-xS isoform and triggering apoptosis106. However, this requires critical discussion since the G4 motif is found on the template strand, meaning the pre-mRNA cannot not form the G4. Probably the G4 creates an obstacle for the RNA polymerase, causing reduced polymerase speed or stalling. Of note, proteins recognizing methylated DNA can slow down RNA polymerase II, thereby indirectly influencing alternative splicing mechanisms107. In line with this, targeted demethylation via TET1 linked to dCas9 causes exon skipping108. Aberrant splicing regulation can result in readthroughs to neighboring genes during stress conditions109, which could explain the overexpressed lncRNA ENSG00000267629 in MAFLD. In summary, the observed differential methylation in MAFLD groups and in HCC, in combination with G4 formation, suggests a complex interplay between transcription and probably also alternative splicing processes (Fig. 5B).
Conclusion
We were able to show aberrant DNA methylation within the investigated intragenic regions of GCK and TM6SF2 in metabolic diseases as well as in HCC and that these structures are alternative regulatory elements potentially controlled by G4 formation. The link between G4s and cancer is well known. While the number of G4s naturally decreases during cell differentiation110, G4s are increased in tumor tissue111 and in the human keratinocytes (HaCaT) cancer cell line versus normal human epidermal keratinocytes (NHEK)112. Even the cell-type specific transcriptome has been shown to be shaped by G4s113. Therefore, G4s harbor a great potential as novel therapeutics and there are already G4 ligands which are used in clinical trial for solid tumors with mutations in genes involved in homologous recombination repair or pancreatic cancers (reviewed in114). Recently it has been shown that pathologically high glucose concentrations cause the formation of a stable promoter-G4 and in turn result in significantly increased gene expression compared to normal glucose concentrations45. We suggest that a similar mechanism also applies for MetD patients since they are exposed constantly to high glucose and high fatty acid concentrations, if not treated properly.
Although further research is necessary to get a deeper understanding of the mechanisms involved, we were able to connect detected DMPs in metabolic risk genes to G-quadruplex formation in a patient cohort suffering from metabolism dysfunction-associated fatty liver disease and type II diabetes, for the very first time. Unfortunately, we are currently lacking G4 formation data in human liver biopsies, since this type of material is rare. Although the assessment of anthropometric and blood parameters, abdominal MRI and liver biopsies are the current standard for MAFLD diagnosis, liver biopsies cannot be applied on a broad scale in asymptomatic obese or lean-MAFLD subjects for ethical reasons or low patient acceptance. In conclusion, since metabolic diseases are linked to an elevated cancer risk, and aberrant DNA methylation as well as elevated non-B DNA formation occurs during cancer formation, these mechanisms could point to a common mechanism of metabolic diseases and liver cancer development. Understanding this intricate connection especially in intragenic regions with alternative regulatory roles is paramount in unraveling the complexities of metabolic diseases, potentially leading to G4s as novel targets for metabolic disease treatment as already evaluated for solid cancers.
Methods
Study design
We designed a nested cohort study by selecting 148 blood samples from Caucasian origin collected at the University Hospital Salzburg, Austria, which fit into one of five groups according to the criteria below. The local ethics committee (Ethikkommission des Landes Salzburg No. 415-E/1521/6–2012, and an ethics approval by the Paracelsus Medical University Salzburg No. 01–2022) approved the study and informed consent was obtained from all participants. The study was conducted in accordance with the guidelines of the World Medical Association’s Declaration of Helsinki.
Alanine transaminase (ALT) and aspartate transaminase (AST) levels ≤ 40 U/L for women and ≤ 50 U/L for men, with or without increased levels of γ-glutamyl transpeptidase (GGT), were considered as normal liver tests. Routine clinical parameters were determined during sampling at the outpatient clinics of the University Hospital Salzburg, Austria.
-
(1)
Lean healthy subjects (LH, n = 24): body mass index (BMI) ≤ 25.0 kg/m2, normal liver tests, normal ultrasound and no components of MetS according to ATPIII criteria115
-
(2)
Obese healthy subjects (OH, n = 31): metabolic healthy obese (MHO) BMI ≥ 30.0 kg/m2, normal ultrasound and normal liver tests, and ≤ 2 components of MetS (obesity and one additional metabolic syndrome defining criterion allowed and glycated hemoglobin (HbA1c) < 6.0%;
-
(3)
Lean MAFLD subjects (LM, n = 27): metabolic unhealthy normal weight (MUNW), BMI ≤ 25.0 kg/m2, ultrasound evidence of a fatty liver, with or without elevated liver tests, HbA1c < 7%;
-
(4)
Obese MAFLD subjects (OM, n = 32): BMIs ≥ 30.0 kg/m2, unequivocal fatty liver ultrasound, with or without elevated liver tests, HbA1c < 7%;
-
(5)
Advanced MetD (aMetD, n = 34): Elevated fibrosis-4 (FIB4) score > 2.67 and/or established T2D with elevated HbA1c > 7%.
Sequence specific DNA methylation analysis by pyrosequencing
DNA was extracted from 200 µL whole blood with the ReliaPrep Blood gDNA Miniprep System (Promega, Austria) according to the manufacturer´s guidelines. Bisulfite (BS) conversion was performed with 500 ng of genomic DNA (gDNA) input material using a Bisulfite Conversion Kit (Zymo Research, Austria). DNA was eluted with 60 µL M-Elution buffer (~ 8 ng/µL). PCR was performed in 25 µL reaction volume containing 200 µM dNTPs, 1 mM MgCl2, each 0.2 µM forward and reverse primer, 0.63 U HotStar Taq polymerase (Qiagen, Austria) and 16 ng (P2RXL1, SCGN and NHLRC1) or 32 ng (EDARADD, IPO8, GCK and TM6SF2) of BS-converted DNA to reach sufficient material for pyrosequencing. The PCR protocol included an initial heating step of 95°C for 15 min, followed by 45 cycles of 95°C for 30 s, 55–64°C primer annealing depending on the target (see Supplement Material & Methods Sect. 1.1) for 30 s and 72°C for 15 s, as well as a final elongation step at 72°C for 7 min. Pyrosequencing was performed using 20 µL PCR product and 7.5 µM (IPO8, GCK, TM6SF2, P2RXL1, and NHLRC1) or 9 µM (SCGN and EDARADD) sequencing primer. Detailed assay design, primer sequences, annealing temperatures and sequencing conditions can be found in the Supplement (Material & Methods Sect. 1.1). Data analysis was performed with Pyromark Q24 Advanced software, R version 4.3.1116 and R studio117.
rs58542926 C > T single nucleotide polymorphism (SNP) genotyping by pyrosequencing
To infer the genotype of the rs58542926 C > T (E167K) risk variant, we amplified the region in 25 µL reaction volume using 200 µM dNTPs, 1 mM MgCl2, each 0.2 µM forward and reverse primer, 0.63 U HotStar Taq polymerase (Qiagen, Austria) and 10 ng genomic DNA. The PCR protocol included an initial heating step of 95°C for 15 min, followed by 45 cycles of 95°C for 30 s, 63°C for 30 s and 72°C for 15 s, as well as a final elongation step at 72°C for 7 min. For the pyrosequencing reaction, we used 20 µl PCR product and 7.5 µM sequencing primer. Detailed information is given in the Supplement Material & Methods Sect. 1.1. Data analysis was performed with Pyromark Q24 Advanced software, R version 4.3.1116 and R studio117.
Gene expression data
Expression data for MAFLD and HCC were extracted from the liver expression portal (GepLiver)72. For the analysis, we downloaded the datasets for Bulk-01 (normal liver), Bulk-05–09 (including MAFLD) and Bulk-19 (TCGA; including HCC) datasets as listed in the GepLiver portal. All statistical analyses were performed using R version 4.3.1116 and R Studio117.
Non-B DNA prediction tools
To predict non-B forming sites, we utilized pqsfinder for G4 forming sites77, QmRLFS-finder for R-loop forming sequences (RLFS)76, and the RNAfold web server of the University of Vienna/Austria for the folding of hairpins118.
Non-B DNA detection with permanganate/S1 nuclease footprinting with direct adapter ligation and sequencing (PDAL-Seq)
PDAL-Seq was performed according to the protocol described in81. Further details can be found in the Supplement (Material & Methods Sect. 1.3). In short, 5–7 × 106 cells were washed two times with 1 × PBS without Ca2+ and Mg2+ and incubated in 1 mL low salt buffer (15 mM Tris–HCl pH 7.5, 15 mM NaCl, 60 mM KCl, 300 mM sucrose, 0.5 mM EGTA) for 1 min. 670 µL of freshly prepared KMnO4 stock solution (40 mM for HEK293, 70 mM for MCF7) were added and incubated at 37°C in the dark for 80 s. Subsequently, the reaction was stopped with 2.67 mL stop solution (50 mM EDTA, 700 mM β-mercaptoethanol, 1% SDS) and 79.5 µL proteinase K (20 mg/ml stock solution, Promega, Austria) were added. After overnight incubation at 37°C, DNA was extracted with phenol chloroform isoamylalcohol (PCI, Merck, Austria) and precipitated with 2 M NH4OAc and 100% ethanol. After resuspending the pellet in 1 mL of 10 mM Tris HCl pH 8.0 buffer, RNA was digested by adding 2.5 µL of 20 mg/mL RNase A (Promega, Austria) for 1 h followed by another PCI extraction. Free DNA ends were blocked with 500 U Terminal Transferase (Roche, Austria), 120 µM cordycepin-5´-triphosphate and 5 mM CoCl2 in 500 µL total reaction volume, followed by a PCI extraction and two ethanol precipitations to avoid cordycepin carry over. 40 µg DNA were digested with 20 U S1 nuclease (0.5 U/1 µg DNA, Promega, Austria) in a total reaction volume of 600 µL at 37°C for 20 min. The reaction was stopped by adding 10 mM EDTA. After PCI extraction, end repair using T4 DNA polymerase (NEB, Austria) was performed for 15 min at 12°C, followed by PCI extraction. 500 nM biotinylated P5 adapter (Merck, Germany) were linked with 2000 U T4 DNA ligase (Promega, Austria) in a final volume of 100 µL at 16 °C overnight. To remove unligated P5 adapters, Agencourt SPRI bead cleanup (Beckman Coulter, Austria) was performed with 0.9 volumes of beads and eluted in 132 µL 1 × TE buffer (10 mM Tris HCl pH 8.0, 1 mM EDTA). Sonication was performed with an Covaris M220 focused-ultrasonicator in 130 µL microTube-AFA Fiber Pre-Slit Snap Cap tubes with the parameters: peak incidence power: 75, duty factor: 10, cycles per burst: 200 and treatment time: 80 s, to reach a target peak of approximately 400 bp. Fragments were bound to streptavidin coated magnetic beads (Dynabeads® M-280 kilobaseBINDER kit, Thermo Scientific, Austria) and carefully washed with 10 mM Tris–HCl pH 7.5. The beads were then resuspended in 100 µL end repair mix (NEBNext End Repair Module, NEB, Austria) and incubated at 20°C for 30 min. After washing with 10 mM Tris–HCl pH 7.5, P7 adapter ligation (1 µM adapters, 400 U T4 DNA ligase) was performed on a wheel tube rotator at 20°C for 4 h. Beads were washed with 10 mM Tris–HCl pH 7.5 and resuspended in 100 µL PCR mix and PCR was performed. Detailed information about PCR reaction and primers are provided in the methods paper81 and in the Supplement (Material and Methods Sect. 1.3). SPRI bead purification removed unbound adapters and primer dimers and the quality of the resulting libraries was checked using an Agilent Fragment Analyzer. Library concentration was determined with a KAPA Library Quantification Kit (Roche, Austria) and sequenced on a NovaSeq platform with approximately 80 million reads. After demultiplexing the raw reads and quality control, reads were aligned to the human reference genome hg19 with the BWA-MEM aligner119. Subsequent filtering removed low-quality reads (mapping quality MQ < 30) and ENCODE blacklisted regions82. BamCoverage files were generated with a 50 bp bin size and normalized to 1 × coverage and visualized in IGV120. Data analysis was performed using the open-source platform Galaxy121.
Studying in vitro G4 formation with native DNA polyacrylamide gels and circular dichroism (CD) spectroscopy
Target regions harboring the DMPs in GCK and TM6SF2, a positive control region in MYC containing a predicted G4 motif122, and a negative control region within a regulatory region of the estrogen receptor substrate 1 (ESR1) without a predicted G4 motif123 were PCR-amplified. To do so, we prepared a 10 µL PCR reaction containing 200 µM dNTPs, 1 mM MgCl2, each 0.2 µM forward and reverse primer, 0.25 U HotStar Taq polymerase (Qiagen, Austria) and 10 ng genomic DNA. The PCR protocol included an initial heating step of 95°C for 15 min, followed by 45 cycles of 95°C for 30 s, 54–63°C primer annealing depending on the gene for 30 s and 72°C for 15 s, as well as a final elongation step at 72°C for 7 min. The PCR fragments were cleaned-up with ReliaPrep DNA concentration kit (Promega, Austria) and eluted with dH2O. The amplicons were then diluted to a final concentration of 20 ng/µL in 50 mM Tris–HCl (pH 8.3). To enhance G4 folding, we prepared the same dilutions and added 150 mM KCl or 150 mM KCl and 10 µM pyridostatin (PDS, Merck, Austria) to the Tris–HCl buffer. The fragments were then heated to 95°C and cooled gradually down to 25°C in a thermocycler overnight. DNA fragments were loaded onto a native 10% DNA polyacrylamide (PAA) gel using 1 × TBE buffer at 68 V (8 V/cm2) for 2h. DNA PAA gels were post-stained with GelRed (Biotium, VWR, Austria) for 5 min and visualized with a ChemiDoc station (BioRad, Austria). Detailed information can be found in the Supplement (Material & Methods Sect. 1.3) as well as the raw gel image in Supplement Fig. 7.
Circular dichroism (CD) spectra were obtained from single stranded DNA oligos from the predicted G4 motif as well as in a methylated and mutated form (Supplement Material & Methods Sect. 1.4). All oligos were reconstituted in 50 mM Tris–HCl (pH 7.5) and ranging from 0 mM, 50 mM to 150 mM KCl in a final volume of 200 µL with an oligo concentration of 5µM. CD spectra were recorded using the Chirascan™ Plus CD Spectrophotometer instrument (Applied Photophysics, United Kingdom) in a 0.5 mm path cuvette at 25°C. Spectra were scanned from 340 to 220 nm, with a step size of 0.2 nm and a bandwidth of 1 nm. Four spectra per sample were averaged. After blank correction, the CD ellipticity θ in millidegrees was normalized using the relation θnorm = θ / (32,980 × c × l), where c is the DNA concentration in mol/L and l is the path length in cm.
TCGA data and ENCODE histone mark and TF binding data analysis
DNA methylation and gene expression data were obtained from tumor versus normal tissue from patients suffering from hepatocellular carcinoma (HCC) via the Shiny Methylation Analysis Resource Tool (SMART), an interactive tool for analyzing The Cancer Genome Atlas (TCGA) data124.
The Encyclopedia of DNA Elements (ENCODE) data for H3K27ac, H3K4me1 and H3K4me3 from the liver hepatoblastoma cell line HepG2 and the lymphoblast cell line K56282 were downloaded and visualized using the Integrative Genome Viewer (IGV)120. Early growth response 1 (EGR1) chromatin immunoprecipitation and sequencing (ChIP-Seq) data from K562 cells were obtained from the ENCODE database82.
Analyzing contact points in HiC datasets
Genome organization and chromatin interaction data (HiC) were obtained from HepG2 cells via ENCODE82, and for HCC material83, and visualized with 3D Genome Browser84. To detect potential interactions with surrounding genes, we used the coordinates chr7: 44,187,355 in hg19 (chr7:44,147,756; hg38) for GCK exon 7 and chr19:19,381,266 in hg19 (chr19:19,270,457; hg38) for TM6SF2 intron 2-exon 3 for virtual 4C analysis using HiC data in the 3D Genome Browser84.
Testing for regulatory functions using the luciferase reporter assay
To verify the regulatory role of the DMPs identified in GCK exon 7 and TM6SF2 intron 2-exon 3 boundary, the regions were cloned into pCpG-free vectors (Invivogen, Belgium). The pCpG-free basic vector was used to detect potential promoter activity and the pCpG-free promoter vector contains a minimal promoter to test for enhancer activity of the regions of interest. Since these vectors do not contain any CpG dinucleotides in the backbone, in vitro DNA methylation can be performed with the M.SssI enzyme, which then selectively methylates only the inserted human GCK or TM6SF2 region and allows to selectively study the effect of 5mC in the insert on luciferase gene expression.
Briefly, DNA was isolated from HepG2 cells and the regions were amplified in 50 µL reaction volumes containing 1 × GC buffer, 200 µM dNTPs, 0.5 µM each forward and reverse primer, and 0.5 U Phusion HSII polymerase (Thermo Scientific, Austria). The PCR protocol contained an initial heating step of 98°C for 30 s, followed by 45 cycles of 98°C for 10 s, 69°C for 10 s and 72°C for 15 s, as well as a final elongation step at 72°C for 5 min. The PCR primers contained restriction enzyme (RE) cutting sites for NsiI (NEB, Austria) and BamHI/HindIII (NEB, Austria) at the 5´ends of the forward and reverse primers, respectively. 500 ng of each, PCR amplicon and plasmid, were double-digested at 37°C for 20 min and cleaned up with a DNA Clean-up and Concentration System (Promega, Austria). Detailed information can be found in the Supplement (Material & Methods Sect. 1.5).
The ligation reaction was performed in a 1:5 plasmid to insert copy number ratio using T4 DNA ligase (NEB, Austria) at 16°C overnight. 5 µL of the ligated construct were transferred to chemically competent E.coli GT115 cells (pir mutant strain, Invivogen, Belgium) and spread on lysogeny broth (LB) plates containing zeocin (50 µL/ml; Invivogen, Belgium). Singe colonies were picked and incubated in 3 ml LB-zeocin medium overnight. Plasmid was extracted with PureYield Plasmid Midiprep System (Promega, Austria) and successful cloning was verified by Sanger Sequencing (LGC genomics, Germany). 1 µg of plasmid was incubated with 8 U M.SssI (NEB, Austria) at 37°C for 4 h for in vitro DNA methylation. The reaction was heat deactivated at 65°C for 20 min, followed by plasmid cleanup with the ReliaPrep DNA Clean-Up and Concentration System (Promega, Austria).
24 h prior to transfection, 0.15 × 105 HepG2 cells per well were seeded in 24-well plates with DMEM low glucose medium + 10% fetal bovine serum without antibiotics. Lipofectamin 3000 (Thermo Scientific, Austria) was used to transfect 500 ng plasmid (renilla luciferase) and 25 ng pGL4 SV40 background plasmid (firefly luciferase) and incubated overnight. After cell lysis, chemiluminescent read-out was detected on a Tecan Spark plate reader (Tecan, Austria). Renilla readouts were normalized to the firefly luciferase and finally, data were expressed as fold-change normalized against the empty vector.
Statistical analyses
All statistical analyses were performed using R version 4.3.1116 and R Studio117. Significant changes in DNA methylation in all groups were detected using analysis of variance (ANOVA) with p < 0.05. For individual group comparisons, Student´s t-test (p < 0.05) was applied. DNA methylation data from TCGA and gene expression data from GepLiver database were analyzed with Wilcoxon rank sum test using Benjamini Hochberg (BH) correction. Welch´s two sample t-tests were performed to calculate significant differences in luciferase reporter assays (p < 0.05).
Data availability
PDAL-Seq data are available at the Sequence Read Archive (SRA) under the BioProject number PRJNA1088738 (https://www.ncbi.nlm.nih.gov/Traces/study/?acc = PRJNA1088738&o = acc_s%3Aa).
Abbreviations
- 5mC:
-
5-Methylcytosine
- BS:
-
Bisulfite conversion
- ChIP-Seq:
-
Chromatin immunoprecipitation and sequencing
- DMP:
-
Differentially methylated position
- DMR:
-
Differentially methylated region
- G4:
-
G-quadruplex
- GCK:
-
Glucokinase
- HiC/4C:
-
Chromatin conformation capture
- MAFLD:
-
Metabolism dysfunction-associated fatty liver disease
- MetD:
-
Metabolic disease
- MetS:
-
Metabolic syndrome
- Non-B DNA:
-
Non-canonical B-DNA structure
- PDAL-Seq:
-
Permanganate/S1 nuclease footprinting with direct adapter ligation
- PQS:
-
Putative G4 forming sequence
- R-loop:
-
RNA:DNA hybrid
- RLFS:
-
R-loop forming sequence
- T2D:
-
Type 2 diabetes mellitus
- TF:
-
Transcription factor
- TM6SF2:
-
Transmembrane 6 superfamily 2
References
WHO. Obesity and overweight, Fact Sheets, https://www.who.int/en/news-room/fact-sheets/detail/obesity-and-overweight, (2020).
Koenen, M., Hill, M. A., Cohen, P. & Sowers, J. R. Obesity, Adipose Tissue and Vascular Dysfunction. Circ. Res. 128, 951–968. https://doi.org/10.1161/CIRCRESAHA.121.318093 (2021).
Collaboration, N. C. D. R. F. Worldwide trends in body-mass index, underweight, overweight, and obesity from 1975 to 2016: a pooled analysis of 2416 population-based measurement studies in 128.9 million children, adolescents, and adults. Lancet 390, 2627–2642. https://doi.org/10.1016/S0140-6736(17)32129-3 (2017).
Samblas, M., Milagro, F. I. & Martinez, A. DNA methylation markers in obesity, metabolic syndrome, and weight loss. Epigenetics 14, 421–444. https://doi.org/10.1080/15592294.2019.1595297 (2019).
Fabbrini, E., Sullivan, S. & Klein, S. Obesity and nonalcoholic fatty liver disease: biochemical, metabolic, and clinical implications. Hepatology 51, 679–689. https://doi.org/10.1002/hep.23280 (2010).
Yamamura, S. et al. MAFLD identifies patients with significant hepatic fibrosis better than NAFLD. Liver Int. 40, 3018–3030. https://doi.org/10.1111/liv.14675 (2020).
Afshin, A., Reitsma, M. B. & Murray, C. J. L. Health Effects of Overweight and Obesity in 195 Countries. N. Engl. J. Med. 377, 1496–1497. https://doi.org/10.1056/NEJMc1710026 (2017).
Chen, Y. et al. Body mass index and risk of gastric cancer: a meta-analysis of a population with more than ten million from 24 prospective studies. Cancer Epidemiol. Biomarkers. Prev. 22, 1395–1408. https://doi.org/10.1158/1055-9965.EPI-13-0042 (2013).
Genkinger, J. M. et al. A pooled analysis of 14 cohort studies of anthropometric factors and pancreatic cancer risk. Int. J. Cancer 129, 1708–1717. https://doi.org/10.1002/ijc.25794 (2011).
Wang, Y., Wang, B., Shen, F., Fan, J. & Cao, H. Body mass index and risk of primary liver cancer: a meta-analysis of prospective studies. Oncologist 17, 1461–1468. https://doi.org/10.1634/theoncologist.2012-0066 (2012).
Ma, Y. et al. Obesity and risk of colorectal cancer: a systematic review of prospective studies. PLoS One https://doi.org/10.1371/journal.pone.0053916 (2013).
Chlebowski, R. T. et al. Breast cancer incidence and mortality by metabolic syndrome and obesity: The Women’s Health Initiative. Cancer https://doi.org/10.1002/cncr.35318 (2024).
Carreras-Torres, R. et al. Obesity, metabolic factors and risk of different histological types of lung cancer: A Mendelian randomization study. PLoS One https://doi.org/10.1371/journal.pone.0177875 (2017).
Li, M. et al. Association of Metabolic Syndrome With Risk of Lung Cancer: A Population-Based Prospective Cohort Study. Chest https://doi.org/10.1016/j.chest.2023.08.003 (2023).
Grasselli, E. et al. Models of non-Alcoholic Fatty Liver Disease and Potential Translational Value: the Effects of 3,5-L-diiodothyronine. Ann. Hepatol. 16, 707–719. https://doi.org/10.5604/01.3001.0010.2713 (2017).
Younossi, Z. et al. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 15, 11–20. https://doi.org/10.1038/nrgastro.2017.109 (2018).
Roden, M. Mechanisms of Disease: hepatic steatosis in type 2 diabetes–pathogenesis and clinical relevance. Nat. Clin. Pract. Endocrinol. Metab. 2, 335–348. https://doi.org/10.1038/ncpendmet0190 (2006).
Gaggini, M. et al. Non-alcoholic fatty liver disease (NAFLD) and its connection with insulin resistance, dyslipidemia, atherosclerosis and coronary heart disease. Nutrients 5, 1544–1560. https://doi.org/10.3390/nu5051544 (2013).
Chalasani, N. et al. The diagnosis and management of non-alcoholic fatty liver disease: practice Guideline by the American Association for the Study of Liver Diseases, American College of Gastroenterology, and the American Gastroenterological Association. Hepatology 55, 2005–2023. https://doi.org/10.1002/hep.25762 (2012).
Ma, J. et al. A peripheral blood DNA methylation signature of hepatic fat reveals a potential causal pathway for nonalcoholic fatty liver disease. Diabetes 68, 1073–1083. https://doi.org/10.2337/DB18-1193 (2019).
Gerhard, G. S. et al. Differentially methylated loci in NAFLD cirrhosis are associated with key signaling pathways. Clin. Epigen. 10, 93. https://doi.org/10.1186/s13148-018-0525-9 (2018).
Ahrens, M. et al. DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab. 18, 296–302. https://doi.org/10.1016/j.cmet.2013.07.004 (2013).
Kirchner, H. et al. Altered DNA methylation of glycolytic and lipogenic genes in liver from obese and type 2 diabetic patients. Mol. Metab. 5, 171–183. https://doi.org/10.1016/j.molmet.2015.12.004 (2016).
Nano, J. et al. Epigenome-wide association study identifies methylation sites associated with liver enzymes and hepatic steatosis. Gastroenterology https://doi.org/10.1053/j.gastro.2017.06.003 (2017).
Rauchhaus, J., Robinson, J., Monti, L. & Di Antonio, M. G-quadruplexes Mark Sites of Methylation Instability Associated with Ageing and Cancer. Genes (Basel) https://doi.org/10.3390/genes13091665 (2022).
Mao, S. Q. et al. DNA G-quadruplex structures mold the DNA methylome. Nat. Struct. Mol. Biol. 25, 951–957. https://doi.org/10.1038/s41594-018-0131-8 (2018).
Williamson, J. R., Raghuraman, M. K. & Cech, T. R. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell 59, 871–880. https://doi.org/10.1016/0092-8674(89)90610-7 (1989).
Sen, D. & Gilbert, W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature 334, 364–366. https://doi.org/10.1038/334364a0 (1988).
Haider, S., Parkinson, G. N. & Neidle, S. Crystal structure of the potassium form of an Oxytricha nova G-quadruplex. J. Mol. Biol. 320, 189–200. https://doi.org/10.1016/S0022-2836(02)00428-X (2002).
Verma, A. et al. Genome-wide computational and expression analyses reveal G-quadruplex DNA motifs as conserved cis-regulatory elements in human and related species. J. Med. Chem. 51, 5641–5649. https://doi.org/10.1021/jm800448a (2008).
Huppert, J. L. & Balasubramanian, S. G-quadruplexes in promoters throughout the human genome. Nucl. Acids Res. 35, 406–413. https://doi.org/10.1093/nar/gkl1057 (2007).
Williams, J. D. et al. Characterization of long G4-rich enhancer-associated genomic regions engaging in a novel loop:loop “G4 Kissing” interaction. Nucl. Acids Res. 48, 5907–5925. https://doi.org/10.1093/nar/gkaa357 (2020).
Georgakopoulos-Soares, I. et al. Alternative splicing modulation by G-quadruplexes. Nat. Commun. 13, 2404. https://doi.org/10.1038/s41467-022-30071-7 (2022).
Guiblet, W. M. et al. Selection and thermostability suggest G-quadruplexes are novel functional elements of the human genome. Genome Res. 31, 1136–1149. https://doi.org/10.1101/gr.269589.120 (2021).
Duquette, M. L., Handa, P., Vincent, J. A., Taylor, A. F. & Maizels, N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 18, 1618–1629. https://doi.org/10.1101/gad.1200804 (2004).
Raiber, E. A., Kranaster, R., Lam, E., Nikan, M. & Balasubramanian, S. A non-canonical DNA structure is a binding motif for the transcription factor SP1 in vitro. Nucl. Acids Res. 40, 1499–1508. https://doi.org/10.1093/nar/gkr882 (2012).
Spiegel, J. et al. G-quadruplexes are transcription factor binding hubs in human chromatin. Genome Biol. 22, 117. https://doi.org/10.1186/s13059-021-02324-z (2021).
Smith, S. S., Kan, J. L., Baker, D. J., Kaplan, B. E. & Dembek, P. Recognition of unusual DNA structures by human DNA (cytosine-5)methyltransferase. J. Mol. Biol. 217, 39–51. https://doi.org/10.1016/0022-2836(91)90609-a (1991).
Chen, X., Mariappan, S. V., Moyzis, R. K., Bradbury, E. M. & Gupta, G. Hairpin induced slippage and hyper-methylation of the fragile X DNA triplets. J. Biomol. Struct. Dyn. 15, 745–756. https://doi.org/10.1080/07391102.1998.10508989 (1998).
Frasson, I., Pirota, V., Richter, S. N. & Doria, F. Multimeric G-quadruplexes: A review on their biological roles and targeting. Int. J. Biol. Macromol. 204, 89–102. https://doi.org/10.1016/j.ijbiomac.2022.01.197 (2022).
Varshney, D., Spiegel, J., Zyner, K., Tannahill, D. & Balasubramanian, S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 21, 459–474. https://doi.org/10.1038/s41580-020-0236-x (2020).
Makova, K. D. & Weissensteiner, M. H. Noncanonical DNA structures are drivers of genome evolution. Trends. Genet. 39, 109–124. https://doi.org/10.1016/j.tig.2022.11.005 (2023).
Fujii, T., Podbevsek, P., Plavec, J. & Sugimoto, N. Effects of metal ions and cosolutes on G-quadruplex topology. J. Inorg. Biochem. 166, 190–198. https://doi.org/10.1016/j.jinorgbio.2016.09.001 (2017).
Nishio, M., Tsukakoshi, K. & Ikebukuro, K. G-quadruplex: Flexible conformational changes by cations, pH, crowding and its applications to biosensing. Biosens. Bioelectron. https://doi.org/10.1016/j.bios.2021.113030 (2021).
Guneri, D. et al. Structural Insights into Regulation of Insulin Expression Involving i-Motif DNA Structures in the Insulin-Linked Polymorphic Region. Biorxiv https://doi.org/10.1101/2023.06.01.543149 (2023).
Perez, A. et al. Impact of methylation on the physical properties of DNA. Biophys. J. 102, 2140–2148. https://doi.org/10.1016/j.bpj.2012.03.056 (2012).
Li, P. T. et al. Expression of the human telomerase reverse transcriptase gene is modulated by quadruplex formation in its first exon due to DNA methylation. J Biol Chem 292, 20859–20870. https://doi.org/10.1074/jbc.M117.808022 (2017).
Lin, J. et al. Stabilization of G-quadruplex DNA by C-5-methyl-cytosine in bcl-2 promoter: implications for epigenetic regulation. Biochem. Biophys. Res. Commun. 433, 368–373. https://doi.org/10.1016/j.bbrc.2012.12.040 (2013).
Yoshida, W. et al. Detection of DNA Methylation of G-Quadruplex and i-Motif-Forming Sequences by Measuring the Initial Elongation Efficiency of Polymerase Chain Reaction. Anal. Chem. 88, 7101–7107. https://doi.org/10.1021/acs.analchem.6b00982 (2016).
Fry, M. & Loeb, L. A. The fragile X syndrome d(CGG)n nucleotide repeats form a stable tetrahelical structure. Proc. Natl. Acad. Sci. U S A 91, 4950–4954. https://doi.org/10.1073/pnas.91.11.4950 (1994).
Stevens, A. J. et al. G-quadruplex structures and CpG methylation cause drop-out of the maternal allele in polymerase chain reaction amplification of the imprinted MEST gene promoter. PLoS One https://doi.org/10.1371/journal.pone.0113955 (2014).
Tsukakoshi, K., Saito, S., Yoshida, W., Goto, S. & Ikebukuro, K. CpG Methylation Changes G-Quadruplex Structures Derived from Gene Promoters and Interaction with VEGF and SP1. Molecules https://doi.org/10.3390/molecules23040944 (2018).
Halder, R. et al. Guanine quadruplex DNA structure restricts methylation of CpG dinucleotides genome-wide. Mol. Biosyst. 6, 2439–2447. https://doi.org/10.1039/c0mb00009d (2010).
Jara-Espejo, M. & Line, S. R. DNA G-quadruplex stability, position and chromatin accessibility are associated with CpG island methylation. FEBS J. 287, 483–495. https://doi.org/10.1111/febs.15065 (2020).
Singer, G. A. et al. Genome-wide analysis of alternative promoters of human genes using a custom promoter tiling array. BMC Genom. 9, 349. https://doi.org/10.1186/1471-2164-9-349 (2008).
Wang, J., Ungar, L. H., Tseng, H. & Hannenhalli, S. MetaProm: a neural network based meta-predictor for alternative human promoter prediction. BMC Genom. 8, 374. https://doi.org/10.1186/1471-2164-8-374 (2007).
Andersson, R. et al. An atlas of active enhancers across human cell types and tissues. Nature 507, 455–461. https://doi.org/10.1038/nature12787 (2014).
Nepal, C. & Andersen, J. B. Alternative promoters in CpG depleted regions are prevalently associated with epigenetic misregulation of liver cancer transcriptomes. Nat. Commun. 14, 2712. https://doi.org/10.1038/s41467-023-38272-4 (2023).
Schamschula, E. et al. Disease-related blood-based differential methylation in cystic fibrosis and its representation in lung cancer revealed a regulatory locus in PKP3 in lung epithelial cells. Epigenetics 17, 837–860. https://doi.org/10.1080/15592294.2021.1959976 (2022).
Hattersley, A. T. et al. Linkage of type 2 diabetes to the glucokinase gene. Lancet 339, 1307–1310. https://doi.org/10.1016/0140-6736(92)91958-b (1992).
Njolstad, P. R. et al. Neonatal diabetes mellitus due to complete glucokinase deficiency. N. Engl. J. Med. 344, 1588–1592. https://doi.org/10.1056/NEJM200105243442104 (2001).
Rose, C. S. et al. A -30G>A polymorphism of the beta-cell-specific glucokinase promoter associates with hyperglycemia in the general population of whites. Diabetes 54, 3026–3031. https://doi.org/10.2337/diabetes.54.10.3026 (2005).
Xu, L. et al. GCK gene-body hypomethylation is associated with the risk of coronary heart disease. Biomed. Res. Int. https://doi.org/10.1155/2014/151723 (2014).
Pan, J. Q., Zhang, Y. Q., Wang, J. H., Xu, P. & Wang, W. lncRNA co-expression network model for the prognostic analysis of acute myeloid leukemia. Int. J. Mol. Med. 39, 663–671. https://doi.org/10.3892/ijmm.2017.2888 (2017).
Zou, W. et al. A metabolism-related 4-lncRNA prognostic signature and corresponding mechanisms in intrahepatic cholangiocarcinoma. BMC Cancer 21, 608. https://doi.org/10.1186/s12885-021-08322-5 (2021).
Kozlitina, J. et al. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 46, 352–356. https://doi.org/10.1038/ng.2901 (2014).
Tang, S. et al. Association of TM6SF2 rs58542926 T/C gene polymorphism with hepatocellular carcinoma: a meta-analysis. BMC Cancer 19, 1128. https://doi.org/10.1186/s12885-019-6173-4 (2019).
Bresgen, N., Kovacs, M., Lahnsteiner, A., Felder, T. K. & Rinnerthaler, M. The Janus-Faced Role of Lipid Droplets in Aging: Insights from the Cellular Perspective. Biomolecules https://doi.org/10.3390/biom13060912 (2023).
ENSEMBL. https://www.ensembl.org/index.html, (2013).
Kent, W. J. et al. The human genome browser at UCSC. Genome Res. 12, 996–1006. https://doi.org/10.1101/gr.229102 (2002).
Cancer Genome Atlas Research, N. et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 45, 1113–1120 https://doi.org/10.1038/ng.2764. (2013).
Li, Z. et al. GepLiver: an integrative liver expression atlas spanning developmental stages and liver disease phases. Sci. Data 10, 376. https://doi.org/10.1038/s41597-023-02257-1 (2023).
Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 14, R115. https://doi.org/10.1186/gb-2013-14-10-r115 (2013).
Hannum, G. et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell 49, 359–367. https://doi.org/10.1016/j.molcel.2012.10.016 (2013).
Levine, M. E. et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY) 10, 573–591. https://doi.org/10.18632/aging.101414 (2018).
Jenjaroenpun, P., Wongsurawat, T., Yenamandra, S. P. & Kuznetsov, V. A. QmRLFS-finder: A model, web server and stand-alone tool for prediction and analysis of R-loop forming sequences. Nucl. Acids Res. 43, 10081. https://doi.org/10.1093/nar/gkv974 (2015).
Hon, J., Martinek, T., Zendulka, J. & Lexa, M. pqsfinder: an exhaustive and imperfection-tolerant search tool for potential quadruplex-forming sequences in R. Bioinformatics 33, 3373–3379. https://doi.org/10.1093/bioinformatics/btx413 (2017).
Marsico, G. et al. Whole genome experimental maps of DNA G-quadruplexes in multiple species. Nucleic Acids Res. 47, 3862–3874. https://doi.org/10.1093/nar/gkz179 (2019).
Li, C. et al. Ligand-induced native G-quadruplex stabilization impairs transcription initiation. Genome Res. 31, 1546–1560. https://doi.org/10.1101/gr.275431.121 (2021).
Kouzine, F. et al. Permanganate/S1 Nuclease Footprinting Reveals Non-B DNA Structures with Regulatory Potential across a Mammalian Genome. Cell Syst. https://doi.org/10.1016/j.cels.2017.01.013 (2017).
Lahnsteiner, A. et al. In vivo detection of DNA secondary structures using Permanganate/S1 Footprinting with Direct Adapter Ligation and Sequencing (PDAL-Seq). Methods Enzymol. https://doi.org/10.1016/bs.mie.2023.12.003 (2024).
ENCODE. ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 https://doi.org/10.1038/nature11247. (2012).
Xu, Z. et al. Structural variants drive context-dependent oncogene activation in cancer. Nature 612, 564–572. https://doi.org/10.1038/s41586-022-05504-4 (2022).
Wang, Y. et al. The 3D Genome Browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol. 19, 151. https://doi.org/10.1186/s13059-018-1519-9 (2018).
Lou, Y. et al. Potential Regulators Driving the Transition in Nonalcoholic Fatty Liver Disease: a Stage-Based View. Cell Physiol. Biochem. 41, 239–251. https://doi.org/10.1159/000456061 (2017).
Zhang, W. et al. Fibroblast-specific adipocyte enhancer binding protein 1 is a potential pathological trigger and prognostic marker for liver fibrosis independent of etiology. J. Dig. Dis. 24, 550–561. https://doi.org/10.1111/1751-2980.13230 (2023).
Ho, C. Y., Lee, J. I., Huang, S. P., Chen, S. C. & Geng, J. H. A Genome-Wide Association Study of Metabolic Syndrome in the Taiwanese Population. Nutrients https://doi.org/10.3390/nu16010077 (2023).
Kubickova, K. N. et al. Predictive role BLVRA mRNA expression in hepatocellular cancer. Ann. Hepatol. 15, 881–887. https://doi.org/10.5604/16652681.1222104 (2016).
Tufan, N. L. et al. Hepatitis Bx antigen stimulates expression of a novel cellular gene, URG4, that promotes hepatocellular growth and survival. Neoplasia 4, 355–368. https://doi.org/10.1038/sj.neo.7900241 (2002).
Dodurga, Y., Secme, M. & Lale Satiroglu-Tufan, N. A novel oncogene URG4/URGCP and its role in cancer. Gene 668, 12–17. https://doi.org/10.1016/j.gene.2018.05.047 (2018).
Okuda, J. et al. Persistent overexpression of phosphoglycerate mutase, a glycolytic enzyme, modifies energy metabolism and reduces stress resistance of heart in mice. PLoS One 8, e72173. https://doi.org/10.1371/journal.pone.0072173 (2013).
Di Costanzo, A. et al. Evaluation of Polygenic Determinants of Non-Alcoholic Fatty Liver Disease (NAFLD) By a Candidate Genes Resequencing Strategy. Sci. Rep. 8, 3702. https://doi.org/10.1038/s41598-018-21939-0 (2018).
Zhou, Y. et al. UPF1 inhibits the hepatocellular carcinoma progression by targeting long non-coding RNA UCA1. Sci. Rep. 9, 6652. https://doi.org/10.1038/s41598-019-43148-z (2019).
Kim, M. J. et al. SUGP1 is a novel regulator of cholesterol metabolism. Hum. Mol. Genet. 25, 3106–3116. https://doi.org/10.1093/hmg/ddw151 (2016).
Loyfer, N. et al. A DNA methylation atlas of normal human cell types. Nature 613, 355–364. https://doi.org/10.1038/s41586-022-05580-6 (2023).
Huo, Z. et al. DNA methylation variability in Alzheimer’s disease. Neurobiol. Aging 76, 35–44. https://doi.org/10.1016/j.neurobiolaging.2018.12.003 (2019).
Faltus, C. et al. Identification of NHLRC1 as a Novel AKT Activator from a Lung Cancer Epigenome-Wide Association Study (EWAS). Int. J. Mol. Sci. https://doi.org/10.3390/ijms231810699 (2022).
Ilieva, M. et al. Systematic Analysis of Long Non-Coding RNA Genes in Nonalcoholic Fatty Liver Disease. Noncoding RNA https://doi.org/10.3390/ncrna8040056 (2022).
Perrin-Cocon, L. et al. A hexokinase isoenzyme switch in human liver cancer cells promotes lipogenesis and enhances innate immunity. Commun. Biol. 4, 217. https://doi.org/10.1038/s42003-021-01749-3 (2021).
Cao, X., Mahendran, R., Guy, G. R. & Tan, Y. H. Detection and characterization of cellular EGR-1 binding to its recognition site. J. Biol. Chem. 268, 16949–16957 (1993).
Brazda, V. et al. The Amino Acid Composition of Quadruplex Binding Proteins Reveals a Shared Motif and Predicts New Potential Quadruplex Interactors. Molecules https://doi.org/10.3390/molecules23092341 (2018).
Sun, Z. et al. EGR1 recruits TET1 to shape the brain methylome during development and upon neuronal activity. Nat. Commun. 10, 3892. https://doi.org/10.1038/s41467-019-11905-3 (2019).
Yadav, S. et al. ERK1/2-EGR1-SRSF10 Axis Mediated Alternative Splicing Plays a Critical Role in Head and Neck Cancer. Front. Cell Dev. Biol https://doi.org/10.3389/fcell.2021.713661 (2021).
Trizzino, M. et al. EGR1 is a gatekeeper of inflammatory enhancers in human macrophages. Sci. Adv. https://doi.org/10.1126/sciadv.aaz8836 (2021).
Guo, Y. et al. Zinc finger transcription factor Egf1 promotes non-alcoholic fatty liver disease. JHEP Rep. https://doi.org/10.1016/j.jhepr.2023.100724 (2023).
Le Senechal, R. et al. Alternative splicing of BCL-x is controlled by RBM25 binding to a G-quadruplex in BCL-x pre-mRNA. Nucl. Acids Res. 51, 11239–11257. https://doi.org/10.1093/nar/gkad772 (2023).
Shukla, S. et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 479, 74–79. https://doi.org/10.1038/nature10442 (2011).
Shayevitch, R., Askayo, D., Keydar, I. & Ast, G. The importance of DNA methylation of exons on alternative splicing. RNA 24, 1351–1362. https://doi.org/10.1261/rna.064865.117 (2018).
Hadar, S., Meller, A., Saida, N. & Shalgi, R. Stress-induced transcriptional readthrough into neighboring genes is linked to intron retention. Iscience https://doi.org/10.1016/j.isci.2022.105543 (2022).
Zyner, K. G. et al. G-quadruplex DNA structures in human stem cells and differentiation. Nat. Commun. 13, 142. https://doi.org/10.1038/s41467-021-27719-1 (2022).
Biffi, G., Tannahill, D., Miller, J., Howat, W. J. & Balasubramanian, S. Elevated levels of G-quadruplex formation in human stomach and liver cancer tissues. PLoS One https://doi.org/10.1371/journal.pone.0102711 (2014).
Hansel-Hertsch, R. et al. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 48, 1267–1272. https://doi.org/10.1038/ng.3662 (2016).
Lago, S. et al. Promoter G-quadruplexes and transcription factors cooperate to shape the cell type-specific transcriptome. Nat. Commun. 12, 3885. https://doi.org/10.1038/s41467-021-24198-2 (2021).
Figueiredo, J., Mergny, J. L. & Cruz, C. G-quadruplex ligands in cancer therapy: Progress, challenges, and clinical perspectives. Life Sci. https://doi.org/10.1016/j.lfs.2024.122481 (2024).
Grundy, S. M. et al. Diagnosis and management of the metabolic syndrome: an American Heart Association/National Heart, Lung, and Blood Institute scientific statement: Executive Summary. Crit. Pathw. Cardiol. 4, 198–203. https://doi.org/10.1097/00132577-200512000-00018 (2005).
Team, R. C. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Vienna, Austria. https://www.R-project.org/ (2023).
Team, R. S. RStudio: Integrated Development for R. RStudio, PBC, Boston, MA. http://www.rstudio.com/ (2020).
Vienna, U. o. RNAfold Web Server. http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi (2024). Accessed on 24 July 2024.
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. https://doi.org/10.1093/bioinformatics/btp324 (2009).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26. https://doi.org/10.1038/nbt.1754 (2011).
Jalili, V. et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2020 update. Nucl. Acids Res. 48, W395–W402. https://doi.org/10.1093/nar/gkaa434 (2020).
Zheng, K. W., Chen, Z., Hao, Y. H. & Tan, Z. Molecular crowding creates an essential environment for the formation of stable G-quadruplexes in long double-stranded DNA. Nucl. Acids Res. 38, 327–338. https://doi.org/10.1093/nar/gkp898 (2010).
Lam, E. Y., Beraldi, D., Tannahill, D. & Balasubramanian, S. G-quadruplex structures are stable and detectable in human genomic DNA. Nat. Commun. 4, 1796. https://doi.org/10.1038/ncomms2792 (2013).
Li, Y., Ge, D. & Lu, C. The SMART App: an interactive web application for comprehensive DNA methylation analysis and visualization. Epig. Chromatin 12, 71. https://doi.org/10.1186/s13072-019-0316-3 (2019).
Acknowledgements
We want to thank the Austrian Diabetes Association (ÖDG) very warmly for their support as well as all patients who contributed to the study. Furthermore, we thank Kateryna Makova and Matthias Weissensteiner for plenty of fruitful discussions on PDAL-Seq and their input on G-quadruplex structures.
Funding
A.L was supported by a Starter Grant awarded by the Austrian Diabetes Association (ÖDG) as well as with the Early Career Grant of the University of Salzburg (Pure ID: 35010166). This research was in part funded by the County of Salzburg, Cancer Cluster Salzburg [grant number 20102-P1601064-FPR]. Open access publication was supported by the Paris Lodron University of Salzburg Publication Fund.
Author information
Authors and Affiliations
Contributions
A. L. established PDAL Seq, designed assays and conceptualized the study; A. L., V. E., A. O., Z. L. and E. S. performed experiments, B. P. and E. A. collected and grouped the samples and gave substantial advice and intellectual input to the study; A. L. wrote the manuscript, A. L., E.A. and A.R. revised and edited the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lahnsteiner, A., Ellmer, V., Oberlercher, A. et al. G-quadruplex forming regions in GCK and TM6SF2 are targets for differential DNA methylation in metabolic disease and hepatocellular carcinoma patients. Sci Rep 14, 20215 (2024). https://doi.org/10.1038/s41598-024-70749-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-70749-0
- Springer Nature Limited