Abstract
Fibroblast growth factor 2 (FGF2) is a crucial factor in odontoblast differentiation and dentin matrix deposition, which facilitates pulpodentin repair and regeneration. Nevertheless, the specific biological function of FGF2 in odontoblastic differentiation remains unclear because it is controlled by complex signalling pathways. This study aimed to investigate the mechanism underlying the effect of FGF2 on osteo/odontogenic differentiation of stem cells from the apical papilla (SCAP). SCAP were pretreated with conditioned media containing FGF2 for 1 week, followed by culturing in induced differentiation medium for another week. RNA sequencing (RNA-seq) combined with quantitative reverse transcription polymerase chain reaction (RT-qPCR) was used to evaluate the pathways affected by FGF2 in SCAP. Osteo/odontogenic differentiation of SCAP was determined using Alizarin red S staining, alkaline phosphatase staining, RT-qPCR, and western blotting. Pretreatment with FGF2 for 1 week increased the osteo/odontogenic differentiation ability of SCAP. RNA-seq and Kyoto Encyclopedia of Genes and Genomes pathway analyses revealed that phosphatidylinositol 3-kinase (PI3K)/AKT signalling is involved in the osteogenic function of FGF2. RT-qPCR results indicated that SCAP expressed FGF receptors, and western blotting showed that p-AKT was reduced in FGF2-pretreated SCAP. The activation of the PI3K/AKT pathway partially reversed the stimulatory effect of FGF2 on osteo/odontogenic differentiation of SCAP. Our findings suggest that pretreatment with FGF2 enhances the osteo/odontogenic differentiation ability of SCAP by inhibiting the PI3K/AKT pathway.
Similar content being viewed by others

Introduction
Stem cells from the apical papilla (SCAP) are a type of mesenchymal stem cells (MSCs) that hold great potential for dental regeneration because of their high proliferation, migration, and dental differentiation abilities1,2,3. However, the number of SCAP is limited because they exist only during root development, before the tooth erupts into the oral cavity4,5. Therefore, for their formative functions, it is important to efficiently induce SCAP to proliferate and differentiate into odontoblast-like cells6.
Fibroblast growth factor 2 (FGF2) plays a crucial role in wound healing and tissue formation7,8,9. At the cellular level, FGF2 regulates various cellular functions by activating FGF receptors (FGFR) on the cell membrane10. This activation leads to the dimerisation and activation of intracellular signalling pathways, such as mitogen-activated protein kinase and phosphoinositide three kinase (PI3K)/AKT, which in turn regulate the proliferation, differentiation, and migration of different types of cells11,12.
However, the role of FGF2 in the biological functions of SCAP has not yet been thoroughly investigated. It was reported that FGF2 maintains the expression of stemness markers such as Oct4, Sox2, and Nanog by activating specific signalling pathways, including the ERK1/2 and PI3K/AKT pathways, and subsequently promotes the self-renewal ability of MSCs13. Although the effect of FGF2 on cell proliferation is well established, its effects on mineralisation and osteogenic differentiation are still being debated. Previous studies have shown that the effect of FGF2 on the differentiation of dental MSCs depends on both the cellular phase and duration of FGF2 exposure14,15. In other words, different durations of FGF treatment or pretreatment may either inhibit or enhance the osteo/odontogenic differentiation of SCAP. Specifically, 1 week of FGF2 pretreatment enhances the osteogenic differentiation ability of dental stem cells at the mRNA level. Utilizing the powerful dentinogenic potential of SCAP to enhance its differentiation into odontoblasts and the formation of new dentin under specific conditions is a crucial step in root development and dentin repair. Therefore, based on the stage-specific effects of FGF2 on SCAP differentiation, this article aims to explore the role and specific mechanisms of FGF2 on the osteo/odontogenic differentiation of SCAP at different stages of cell maturation. This provides certain research insights for the application of FGF2 in promoting pulp/dentin regeneration and root formation in SCAP.
Results
Isolation and characterisation of SCAP
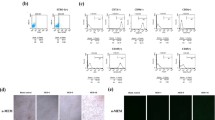
SCAP (Fig. 1A) were capable of forming adherent clonogenic cell clusters, and most of the SCAP retained a fibroblastic spindle shape (Fig. 1B,C). The cells expressed CD24 (Fig. 1D), vimentin (Fig. 1E), and STRO-1 (Fig. 1F), which are positive phenotypic markers of SCAP, whereas keratin was negative (Fig. 1G). The SCAP were robustly Alizarin red S (ARS)-positive (Fig. 1H) and some developed into ORO–positive lipid-laden fat cells (Fig. 1I).
Immunophenotypic characterisation and multiple differentiation analysis of SCAP. (A) Apical papilla from an extracted human third molar with developing root tissues. (B) Single fibroblastic cell attached one day after cell seeding. (C) SCAP grown in colonies. (D) Immunocytofluorescence staining of CD24 (green). (E) Immunocytofluorescence staining of vimentin (red). (F) Immunocytofluorescence staining of STRO-1 (green). (G) Keratin immunofluorescence staining (nuclear DAPI staining was blue). (H) ARS staining for mineralised nodules after SCAP cultured in an osteogenic medium for 3 weeks. (I) Positive staining for lipid droplets with ORO after adipogenic induction for 3 weeks. ARS Alizarin red S staining, SCAP stem cells from the apical papilla, ORO Oil Red O.
Effect of FGF2 on osteo/odontogenic differentiation of SCAP
To investigate the effect of FGF2 on the osteo/odontogenic differentiation ability of SCAP, we analysed the expression of osteo/odontogenic-related genes in both the control and experimental groups using RT-qPCR. After 1 week of growth in the induction medium, the mRNA expression levels of dentin sialophosphoprotein (DSPP), dentin matrix acid phosphoprotein 1 (DMP1), and bone gamma-carboxyglutamate protein (BGLAP) in SCAP were significantly upregulated compared to those in the controls (Fig. 2A). Western blotting analysis (Fig. 2B) revealed that the pretreatment of SCAP with FGF2 significantly increased the expression of DSPP (Fig. 2C), DMP1 (Fig. 2D), and BGLAP (Fig. 2E) at the protein level. ARS staining (Fig. 2F) and quantitative analyses (Fig. 2G) performed to investigate the effects of FGF2 on the cytodifferentiation of SCAP showed that FGF2 pretreatment resulted in a significant increase in the formation of mineralised nodules compared to the controls. Similarly, alkaline phosphatase (ALP) staining (Fig. 2H) and ALP activity analysis (Fig. 2I) demonstrated that FGF2 pretreatment effectively induced osteogenesis.
Impact of FGF2 pretreatment on osteo/odontogenesis differentiation of SCAP. SCAP were pretreated with FGF2 for 1 week in a standard medium. (A,B) FGF2 pretreatment enhanced mRNA and protein expression of DSPP, DMP1, and BGLAP. (C–E) Representative quantitative analysis of DSPP, DMP1, BGLAP protein (***P < 0.001). (F) FGF2 pretreatment enhanced ARS mineralisation. Scale bar = 200 μm. (G) Quantitative measurement of ARS staining (***P < 0.001). (H) FGF2 pretreatment enhanced ALP activity. Scale bar = 200 μm. (I) Quantitative analysis of ALP activity (***P < 0.001). ALP alkaline phosphatase staining, ARS Alizarin red S staining, FGF2 fibroblast growth factor 2, SCAP stem cells from the apical papilla.
RNA-seq results of SCAP pretreated with FGF2
To investigate the mechanism by which FGF2 promotes the osteo/odontogenic differentiation of SCAP, we conducted RNA-seq on SCAP that were either pretreated with FGF2 or left untreated. Heatmap analysis demonstrated 681 DEGs between the two groups, of which 264 mRNA were upregulated and 417 mRNAs were downregulated (Fig. 3A). A volcano plot was used to identify DEGs based on their fold change and P-values (Fig. 3B). Subsequently, we selected several groups with higher fold-changes and validated the results using RT-qPCR (Fig. 3C), which confirmed the reliability of the RNA-seq results. The RT-qPCR results were consistent with the sequencing results. Furthermore, the upregulated genes are related to the PI3K-AKT signalling pathway. For example, VEGFD can activate the PI3K-AKT signalling pathway, promoting cell survival, proliferation, and angiogenesis. VCAN (Versican) can activate the PI3K-AKT signalling pathway by interacting with cell surface receptors such as CD44, promoting cell migration and survival. Gene ontology analysis showed that cellular components were mainly enriched in the plasma membrane, extracellular space, and extracellular regions (Fig. 3D). Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis (Fig. 3E) revealed that the PI3K/AKT pathway was significantly enriched.
PI3K/AKT pathway is involved in FGF2 stimulation of SCAP osteo/odontogenesis differentiation. SCAP were pretreated with either FGF2 and/or SC79 in the standard medium. (A) Clustered heatmap of RNA-seq, with columns representing different transcripts, and rows representing three biological replicates of FGF2-pretreated and untreated SCAP samples. (B) Volcano plot results of DEGs by each exposure groups. (C) RT-qPCR verified the differentially expressed PI3K/AKT pathway genes in the RNA-seq results (**P < 0.01, ***P < 0.001). (D) Gene Ontology enrichment analysis of differentially expressed mRNAs between FGF2 pretreated and untreated SCAP. (E) KEGG analysis of the signalling pathways involved in FGF2 pretreated and activated SCAP. FGF2 fibroblast growth factor 2, KEGG Kyoto Encyclopedia of Genes and Genomes, PI3K phosphoinositide three kinase, RNA-seq RNA sequencing, RT-qPCR quantitative reverse transcription polymerase chain reaction, SCAP stem cells from the apical papilla, DEG differentially expression gene.
FGF2 suppresses PI3K/AKT signalling and drive SCAP mineralisation
SCAP expressed FGFR1 and FGFR2, as demonstrated by RT-qPCR (Fig. 4A). To investigate the relationship between FGF2 pretreatment and the PI3K/AKT signalling pathway, we assessed the changes in AKT and phosphorylated AKT levels using western blotting analysis (Fig. 4B). Based on these results, we determined the appropriate concentration and stimulation time for SC79, a PI3K activator (Supplementary file 6)16. The results revealed that FGF2 pretreatment reduced the phosphorylation levels of AKT and that that this was partially reversed by SC79 (Fig. 4C), indicating that FGF2 pretreatment inhibits the phosphorylation of AKT and potentially affect the biological function of SCAP. We further explored the role of PI3K/AKT signalling in promoting the osteo/odontogenic differentiation of SCAP by FGF2. We added SC79 to the experimental group and assessed the mRNA and protein expression of DSPP, DMP1, and BGLAP using RT-qPCR (Fig. 4D) and western blotting (Fig. 4E), respectively. These results demonstrated that SC79 partially reversed the differentiation-promoting effect of FGF2 pretreatment on SCAP by downregulating the mRNA and protein expression of these markers (Fig. 4D,F). The results of ARS and ALP staining were consistent with the above findings (Fig. 4G–J), indicating that activation of the PI3K/AKT pathway can reduce the mineralisation in SCAP caused by FGF2 pretreatment.
FGF2 pretreatment promoted SCAP osteo/odontogenesis differentiation by the suppression of the PI3K/AKT signalling pathway. SCAP were pretreated with either FGF2 and/or SC79 in the standard medium. (A) FGF2 enhanced FGFR1 and FGFR2 expression (*P < 0.05). (B) AKT and p-AKT protein levels in SCAP pretreated with either FGF2 and/or SCA79. (C) Quantitative analysis of P-AKT/AKT in each groups (*P < 0.05, ***P < 0.001). (D) DSPP, DMP1, and BGLAP mRNA levels in SCAP were measured after subculture in induction medium for 1 week (*P < 0.05, ***P < 0.001). (E) DSPP, DMP1, and BGLAP protein levels in SCAP were measured after subculture in induction medium for 2 weeks. (F) Representative quantitative analysis of DSPP, DMP1, and BGLAP in each group (*P < 0.05, ***P < 0.001). (G) ARS staining in SCAP was measured after subculture in induction medium for 3 weeks. Scale bar = 200 μm. (H) Quantitative analysis of ARS staining (*P < 0.05). (I) ALP staining of SCAP was measured after subculture in induction medium for 1 week. Scale bar = 200 μm. (J) Quantitative analysis of ALP activity (*P < 0.05). ALP alkaline phosphatase staining, ARS Alizarin red S staining, FGF2 fibroblast growth factor 2, FGFR FGF receptor, PI3K phosphoinositide three kinase, RNA-seq RNA sequencing, SCAP stem cells from the apical papilla.
Discussion
Previous studies have indicated that the nature of FGF2 action is complex, and that the role of FGF2 in regulating cell differentiation and dentinogenesis remains controversial17. FGF2 has both positive and negative regulatory effects on osteogenic differentiation18. The biological effects of FGF2 depend on the differentiation stage of osteoblasts and the length and mode of exposure to certain factors19. Previous studies have found that short-term exposure to FGF2 during the early proliferative stage of SCAP growth results in increased mRNA expression of odontogenic markers14,20. Osathanon reported that the addition of FGF2 during osteogenic induction will weaken osteogenic differentiation and enhance the stemness of dental pulp stem cells21. Based on our previous experiments, we changed the treatment conditions and used FGF2 as a pretreatment before osteogenic induction, and discovered that there was a significant increase in the formation of mineralised nodules, an increase in ALP levels, and an upregulation of both the mRNA and protein levels of BGLAP, DMP1, and DSPP in SCAP. These findings further confirmed that FGF2 plays a crucial role in the early differentiation of SCAP into functional odontoblasts. We believe that the pretreatment period with FGF2 in this study could be considered as the preliminary stage of tooth formation. During this stage, there was little induction of the components of odontogenic differentiation in the cell matrix. The function of FGF2 in this period is to promote cell proliferation and migration22. Additionally, FGF2 is embedded in the dentin matrix or stored as a latent complex in the extracellular matrix of the dental pulp, where it can be released and impact cellular events during pulp regeneration or repair23.
The FGF/FGFR pathway is a receptor tyrosine kinase signalling pathway that affects the differentiation of various cells11,24. FGF/FGFR is reported to be associated with the RAS-MAPK, PI3K-AKT, PLCγ, and JAK-STAT signalling pathways, which regulate physiological processes including cell proliferation, differentiation, and survival25,26. For instance, FGF2 can bind to FGFR and activate the RAGE signalling pathway to affect human dermal microvascular endothelial cell angiogenesis26. FGF2 induces neuronal differentiation of canine bone marrow stromal cells via the FGFR2/PI3K/AKT signalling pathway27. Previous studies pointed out that the level of FGFRs increased during early odontoblast differentiation28. This altered expression of FGFRs and their downstream signals may lead to different effects of FGF2 on cell differentiation29. In the present study, FGF2 promoted the expression of FGFR1 and FGFR2 in SCAP. FGF2 activated FGFR1 significantly more strongly than FGFR2, whereas FGFR3 and FGFR4 were barely activated. These results imply that FGFR1 is the primary receptor for FGF2 involved in regulating the biological functions of SCAP. However, although this study demonstrated that FGF2 significantly promoted the expression of FGFR1 in SCAP, further studies are needed to explore the differential activation of various FGFRs by FGF2 in SCAP and its downstream effects on cellular and functional activities.
To further identify the mechanism of FGF2 in the regulation of SCAP biological function, RNA-seq was performed on 3214 DEGs between the experimental and control groups by transcriptome sequencing, and it was found that these genes were related to multiple signalling pathways, including PI3K/AKT, WNT, TNF, and P53. Most of these genes were related to the PI3K/AKT signalling pathway. AKT, one of the non-canonical signals underlying TGFβ/TBR, is a PI3K-dependent serine/threonine kinase that plays specific roles in regulating cellular metabolism and differentiation of various cell types30. Many downstream genes for PI3K/AKT, such as Runx2, ALP, and GSK3, are reported to be associated with osteogenic differentiation31. Tanaka found that the PI3K/AKT pathway regulates the osteo/odontogenic ability of SCAP, which is consistent with the results of our study30. Yosuke revealed that pretreatment with AKT siRNA, LY294402 (specific inhibitors for PI3K), and rapamycin (specific inhibitors for mTOR) enhances the in vitro and in vivo calcified tissue-forming capacity of SCAP, suggesting that suppressive regulation of the PI3K/AKT/mTOR signalling pathway can promote the osteogenic/dentinogenic capacity of SCAP30. We further explored the role of the PI3K/AKT signalling pathway in FGF2-promoted osteo/odontogenic SCAP. RT-qPCR and western blotting analyses showed that SC79 partially reversed the upregulation of DPSS, DMP1, and BGLAP expression caused by FGF2 pretreatment. Furthermore, the ARS and ALP analysis results confirmed that SC79 reversed the enhanced mineralisation effect caused by FGF2 pretreatment. These results indicate that PI3K/AKT is related to FGF2 regulation of osteo/odontogenic differentiation of SCAP, and the suppression of this pathway can re-induce osteo/odontogenic differentiation. These results can be attributed to the fact that FGF2 pretreatment could regulate several signalling pathways, including PI3K/AKT, WNT, TNF, and P53, whereas SC79 is specific only for the PI3K/AKT signalling pathway. Hence, the precise mechanism underlying the effect of FGF2 pretreatment on the osteo/odontogenic differentiation of SCAP requires further investigation.
There are some limitations in this study. A more comprehensive exploration is needed into the signalling pathways involved and their interactions, including examining the intensity and nature of the signal in the nucleus and the crosstalk among signalling pathways, such as WNT, TNF, and P53 and other signaling pathways that show significant differences in our RNA-Seq results. Furthermore, additional validation of our findings through gene-level FGF2 overexpression and in vivo experiments is required.
Based on the stage-specific effects of FGF2 on SCAP differentiation, the results of our study indicate that 1-week pretreatment of FGF2 has the potential to enhance the osteo/odontogenic differentiation of SCAP by activating PI3K/AKT signalling pathways. This helps us to deeply understand the regulation details of FGF2 in stemness and osteo/odontogenic differentiation. Moreover, the regeneration of pulp-dentin by SCAP in vivo is a complex process. It is crucial to maintain the stemness and proliferation capacity of SCAP, and then promote its osteogenic/odontogenic differentiation. Therefore, the complex regulatory role of FGF2 on stem cells plays an important role in this process. Gaining a deep understanding FGF2 function in SCAP in specific settings are necessary, specially the development of FGF2-loaded materials, thereby providing certain guidance for the application of FGF2 in pulp-dentin regeneration.
Methods
Cell isolation and culture
SCAP samples were collected from healthy immature third molars (n = 10) of 5 male and 5 female patients aged 16–22 years. This study was approved by the Ethics Committee of the Hospital of Stomatology, Zunyi Medical University (ethics approval reference number: YJSKTLS-2019-2021-0061). Written informed consent was obtained from all patients prior to the study. The apical papillae were isolated as previously described32,33. Apical papillary tissue was gently separated, cut into small pieces, and digested with 3 mg/mL type I collagenase (Gibco, Grand Island, NY, USA) and 4 mg/mL trypsin (HyClone, Logan, OH, USA) for 40 min. The digested tissue was then filtered through 70-mm strainers and resuspended in standard culture medium. The standard culture medium used was α-MEM (Gibco) supplemented with 10% foetal bovine serum (Gibco) and 1% penicillin–streptomycin (HyClone). SCAP were purified through the limiting dilution method when they reached approximately 80% confluency. All experiments were performed at passages 3–5.
To evaluate the multipotent differentiation capacity of SCAP, the cells were treated with an osteogenic or adipogenic induction medium (Cyagen, Guangzhou, China) for 3 weeks and then stained with ARS for osteoblasts or Oil Red O (ORO) for adipocytes.
Immunophenotypic characterisation
Briefly, the SCAP were fixed with 4% paraformaldehyde for 30 min, permeabilised with 0.5% Triton X-100 for 15 min, and blocked with 10% BSA for 1 h at 25 °C. Primary antibodies (R&D Systems, Emeryville, CA, USA) against CD24, vimentin, STRO-1, and keratin were added, and the SCAP were incubated overnight at 4 °C. The SCAP were then incubated with secondary antibodies (PE-labelled IgM, FITC-labelled IgG Biosynthesis, Beijing, China) for 1 h in the dark. Finally, DAPI (Beyotime, Shanghai, China) was used to stain the nuclei.
Cell counting kit-8 assay
SCAP were seeded into 96-well plates with 10,000 cells per well. Next day, the SC79 groups were treated with 0.9, 1.82, 3.64, 7.28 µg/mL of the AKT activator SC-79 (Selleck, Houston, TX, USA) for 1 week. After 1 week, SCAP were washed with PBS. Each well was added with 100 μL of culture medium containing 10 μL of CCK-8 solution (Beyotime), and then incubated at 37 °C for 2 h. Use a microplate reader (Molecular Devices, California, USA) to measure the absorbance (OD450nm) and calculate the cell survival rate.
Fibroblastic growth factor 2 and SC79 treatment
SCAP in the experimental group were treated with 5 ng/mL FGF2 (Calbiochem, San Diego, CA, USA) for 7 days as earlier described14; however, the control group was grown in the culture medium. After 7 days, the SCAP were digested and seeded into a standard medium. When the cell density of each group reached 70–80% confluency, the medium was changed to an osteogenic medium. The SC79 group were treated with 1.82 µg/mL of the AKT activator SC-79 (Selleck) for 1 week before osteogenic medium treatment as earlier describe34.
Quantitative real-time polymerase chain reaction (RT-qPCR)
The expression levels of osteo/odontogenic differentiation markers, including DSPP, DMP1, and BGLAP, and differentially expressed genes (DEGs) from RNA-seq analysis, such as FN1, DKK1, VEGFD, IGTA4, and VCAN, were examined. Total RNA was extracted with Trizol (Takara, Kyoto, Japan) and transcribed into cDNA using an RNA reverse transcription kit (Takara). Synthesized cDNA was subjected to quantitative real-time polymerase chain reaction. The subsequent PCR amplification step was performed by using SYBR Premix Ex Taq II (Takara). The reaction was performed by using an ABI PRISM 7500 Real-Time PCR system (Applied Biosystems, Benicia, CA) and the following thermal cycling conditions: 40 cycles of 95 °C for 2 min, 95 °C for 15 s, 60 °C for 1 min, and 72 °C for 25 s. The primer sequences are listed in Supplementary S1.
ARS staining
After 3 weeks, the cells were fixed with 4% polyoxymethylene for 20 min, washed thrice with PBS for 5 min, and stained with 0.5% Alizarin Red S (Beyotime) for 20 min. The formation of mineralized nodules was observed under a microscope. Quantitative data were obtained by solubilizing the samples with 10% cetylpyridinium chloride solution (Sigma, Darmstadt, Germany) and measuring their optical density at 490 nm.
ALP staining
The BCIP/NBT Alkaline Phosphatase Colour Development Kit (Beyotime) was used for ALP staining. SCAP were cultured in osteogenic medium for 1 week, and then fixed with 4% paraformaldehyde, and washed wish PBS. The BCIP/NBT staining solution was prepared according to the instructions and then was used to stain the SCAP in dark for 20 min. Subsequently, distilled water was used to terminate the reaction.
RNA sequencing and bioinformatic analyses
Total RNA quantity and purity were analysed using a Bioanalyzer 2100 and RNA 1000 Nano Lab Chip Kit (Agilent, Santa Clara, CA, USA) with RNA integrity number > 7.0. Poly(A) RNA was purified from total RNA (5 μg) using poly-T oligo-attached magnetic beads using two rounds of purification. Following purification, mRNA was fragmented into small pieces using divalent cations. The cleaved RNA fragments were then reverse-transcribed to create a final cDNA library, according to the protocol for the RNA-seq sample preparation kit (Illumina, San Diego, USA). The average insert size for the paired-end libraries was 300 bp (± 50 bp). We then performed paired-end sequencing on an IlluminaHiseq4000 (LC Sciences, LLC, Houston, TX, USA), according to the manufacturer’s instructions. DEGs were identified as previously described34. Briefly, DESeq2/EdgeR with Q value ≤ 0.05 was used for the differential expression analysis of RNA-seq data in terms of |log2FC| > 1 and Q value ≤ 0.05 (DESeq2 or EdgeR) or Q value ≤ 0.001 (DEGseq).
Western blotting analysis
SCAP were lysed using radio-immunoprecipitation assay (RIPA) buffer (Beyotime), and total protein was extracted and normalized according to the concentrations determined by Enhanced BCA Protein Assay Kit (Beyotime). The total protein was then re-suspended with 5× loading buffer (Beyotime) and boiled for 10 min. Twenty-five mg protein from each sample was separated by electrophoresis on 10% polyacrylamide gels (Epizyme, Shanghai, China) and transferred onto polyvinylidene difluoride membranes (PVDF). The blots were blocked with blocking buffer (Beyotime) and incubated with primary antibodies anti-DSPP (Abcam, Cambridge, MA, UK), anti-DMP1 (Abcam), anti-BGLAP (Abcam), anti-p-AKT (Cell Signalling Technology, Boston, MA, USA), and anti-AKT (Cell Signalling Technology) at 4 °C overnight. Afterward, PVDF membranes were incubated with corresponding secondary antibodies for one hour, the bands’ intensity was visualized using the enhanced chemiluminescence (ECL) luminescence reagent (Meilunbio, Dalian, China) and Gel Image System (Tanon, Shanghai, China).
Statistical analysis
Data are presented as mean ± standard deviation from at least three independent experiments. Data were analysed using GraphPad Prism 8 (GraphPad Software, Inc., La Jolla, CA, USA). Differences among groups were compared using one-way analysis of variance or Student’s t-test for comparisons between groups. Statistical significance was set at P < 0.05.
Ethics declaration
This study was approved by Ethics Committee of the Hospital of Stomatology, Zunyi Medical University, with ethics approval reference [YJSKTLS-2019-2021-0061]. Written informed consent was obtained from the patients.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and supplementary materials. More data will be made available (Jiayuan Wu, Email address: wujiayuan@zmu.edu.cn) on reasonable request.
References
Bojic, S. et al. Dental stem cells—Characteristics and potential. Histol. Histopathol. 29(6), 699–706 (2014).
Kang, J. et al. Stem cells from the apical papilla: A promising source for stem cell-based therapy. Biomed Res. Int. 2019, 6104738 (2019).
Huang, G. T. et al. The hidden treasure in apical papilla: The potential role in pulp/dentin regeneration and bioroot engineering. J. Endod. 34(6), 645–651 (2008).
Zheng, C. et al. Stem cell-based bone and dental regeneration: A view of microenvironmental modulation. Int. J. Oral Sci. 11(3), 23 (2019).
Min, J. H. et al. Dentinogenic potential of human adult dental pulp cells during the extended primary culture. Hum. Cell 24(1), 43–50 (2011).
Volponi, A. A., Pang, Y. & Sharpe, P. T. Stem cell-based biological tooth repair and regeneration. Trends Cell Biol. 20(12), 715–722 (2010).
Shi, H. X. et al. The anti-scar effects of basic fibroblast growth factor on the wound repair in vitro and in vivo. PLoS One 8(4), e59966 (2013).
Coffin, J. D., Homer-Bouthiette, C. & Hurley, M. M. Fibroblast growth factor 2 and its receptors in bone biology and disease. J. Endocr. Soc. 2(7), 657–671 (2018).
Komura, M. et al. Tracheal cartilage growth by intratracheal injection of basic fibroblast growth factor. J. Pediatr. Surg. 52(2), 235–238 (2017).
Xuan, Y. H. et al. High-glucose inhibits human fibroblast cell migration in wound healing via repression of bFGF-regulating JNK phosphorylation. PLoS One 9(9), e108182 (2014).
Nugent, M. A. & Iozzo, R. V. Fibroblast growth factor-2. Int. J. Biochem. Cell Biol. 32(2), 115–120 (2000).
Laestander, C. & Engstrom, W. Role of fibroblast growth factors in elicitation of cell responses. Cell Prolif. 47(1), 3–11 (2014).
Lim, W. et al. Fibroblast growth factor 2 induces proliferation and distribution of G(2)/M phase of bovine endometrial cells involving activation of PI3K/AKT and MAPK cell signaling and prevention of effects of ER stress. J. Cell Physiol. 233(4), 3295–3305 (2018).
Qian, J. et al. Basic fibroblastic growth factor affects the osteogenic differentiation of dental pulp stem cells in a treatment-dependent manner. Int. Endod. J. 48(7), 690–700 (2015).
Wu, J. et al. Basic fibroblast growth factor enhances stemness of human stem cells from the apical papilla. J. Endod. 38(5), 614–622 (2012).
Zeng, J. et al. A novel hypoxic lncRNA, HRL-SC, promotes the proliferation and migration of human dental pulp stem cells through the PI3K/AKT signaling pathway. Stem Cell Res. Ther. 13(1), 286 (2022).
Liu, K. et al. The regenerative potential of bFGF in dental pulp repair and regeneration. Front. Pharmacol. 12, 680209 (2021).
Nowwarote, N. et al. Review of the role of basic fibroblast growth factor in dental tissue-derived mesenchymal stemcells. Asian Biomed. 9(3), 271–283 (2015).
Fakhry, A. et al. Effects of FGF-2/-9 in calvarial bone cell cultures: Differentiation stage-dependent mitogenic effect, inverse regulation of BMP-2 and noggin, and enhancement of osteogenic potential. Bone 36(2), 254–266 (2005).
Sagomonyants, K. & Mina, M. Stage-specific effects of fibroblast growth factor 2 on the differentiation of dental pulp cells. Cells Tissues Organs 199(5–6), 311–328 (2014).
Osathanon, T., Nowwarote, N. & Pavasant, P. Basic fibroblast growth factor inhibits mineralization but induces neuronal differentiation by human dental pulp stem cells through a FGFR and PLCgamma signaling pathway. J. Cell Biochem. 112(7), 1807–1816 (2011).
Sagomonyants, K. et al. Enhanced dentinogenesis of pulp progenitors by early exposure to FGF2. J. Dent. Res. 94(11), 1582–1590 (2015).
Galler, K. M. et al. Influence of root canal disinfectants on growth factor release from dentin. J. Endod. 41(3), 363–368 (2015).
Vaccarino, F. M. & Smith, K. M. Increased brain size in autism—What it will take to solve a mystery. Biol. Psychiatry 66(4), 313–315 (2009).
Turner, N. & Grose, R. Fibroblast growth factor signalling: From development to cancer. Nat. Rev. Cancer 10(2), 116–129 (2010).
Helsten, T., Schwaederle, M. & Kurzrock, R. Fibroblast growth factor receptor signaling in hereditary and neoplastic disease: Biologic and clinical implications. Cancer Metastasis Rev. 34(3), 479–496 (2015).
Nakano, R. et al. Fibroblast growth factor receptor-2 contributes to the basic fibroblast growth factor-induced neuronal differentiation in canine bone marrow stromal cells via phosphoinositide 3-kinase/Akt signaling pathway. PLoS One 10(11), e0141581 (2015).
Chang, M. C. et al. Effect of bFGF on the growth and matrix turnover of stem cells from human apical papilla: Role of MEK/ERK signaling. J. Formos Med. Assoc. 119(11), 1666–1672 (2020).
Kim, Y. S. et al. Effects of fibroblast growth factor-2 on the expression and regulation of chemokines in human dental pulp cells. J. Endod. 36(11), 1824–1830 (2010).
Tanaka, Y. et al. Suppression of AKT-mTOR signal pathway enhances osteogenic/dentinogenic capacity of stem cells from apical papilla. Stem Cell Res. Ther. 9(1), 334 (2018).
Huang, M. et al. m6A methylation regulates osteoblastic differentiation and bone remodeling. Front. Cell Dev. Biol. 9, 783322 (2021).
Wu, J. et al. Conditioned medium from periapical follicle cells induces the odontogenic differentiation of stem cells from the apical papilla in vitro. J. Endod. 39(8), 1015–1022 (2013).
Liang, J. et al. A pilot study on biological characteristics of human CD24(+) stem cells from the apical papilla. J. Dent. Sci. 17(1), 264–275 (2022).
Park, S. Y. et al. Inactivation of PI3K/Akt promotes the odontoblastic differentiation and suppresses the stemness with autophagic flux in dental pulp cells. J. Dent. Sci. 17(1), 145–154 (2022).
Funding
This study was supported by grants from Guizhou Provincial Health Commission Science and Technology Fund Project (No. gzwjkj2020-1-163), Science and Technology Plan Project of Guizhou Province (QKH- Foundation-ZK (2022) General 638), The Etiology and Prevention Team Project of Oral Infectious and Malignant Diseases in the Affiliated Stomatological Hospital of Zunyi Medical University (No. (2020)293).
Author information
Authors and Affiliations
Contributions
Zijie Wang: Conceptualisation, methodology, formal analysis, data curation, writing—original draft, writing—review and editing, visualisation. Chuying Chen: Writing—original draft, writing—review and editing, formal analysis. Liying Sun: Acquisition of data, or analysis and interpretation of data. Mei He: Acquisition of data, or analysis and interpretation of data. Ting Huang: Acquisition of data, or analysis and interpretation of data. Jiji Zheng: Acquisition of data, or analysis and interpretation of data. Jiayuan Wu: Conceptualisation, methodology, formal analysis. The final manuscript was reviewed and approved by all the authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, Z., Chen, C., Sun, L. et al. Fibroblast growth factor 2 promotes osteo/odontogenic differentiation in stem cells from the apical papilla by inhibiting PI3K/AKT pathway. Sci Rep 14, 19354 (2024). https://doi.org/10.1038/s41598-024-70123-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-70123-0
- Springer Nature Limited