Abstract
Ulcerative colitis (UC) is a chronic inflammatory disorder of the colon, and its pathogenesis remains unclear. Polyamine metabolic enzymes play a crucial role in UC. In this study, we aimed to identify pivotal polyamine-related genes (PRGs) and explore the underlying mechanism between PRGs and the disease status and therapeutic response of UC. We analyzed mRNA-sequencing data and clinical information of UC patients from the GEO database and identified NNMT, PTGS2, TRIM22, TGM2, and PPARG as key PRGs associated with active UC using differential expression analysis and weighted gene co-expression network analysis (WCGNA). Receiver operator characteristic curve (ROC) analysis confirmed the accuracy of these key genes in UC and colitis-associated colon cancer (CAC) diagnosis, and we validated their relationship with therapeutic response in external verification sets. Additionally, single-cell analysis revealed that the key PRGs were specific to certain immune cell types, emphasizing the vital role of intestinal tissue stem cells in active UC. The results were validated in vitro and in vivo experiments, including the colitis mice model and CAC mice model. In conclusion, these key PRGs effectively predict the progression of UC patients and could serve as new pharmacological biomarkers for the therapeutic response of UC.
Similar content being viewed by others
Introduction
Ulcerative colitis (UC) is a chronic inflammatory disorder that affects the colon and results from a complex interplay of various factors related to the immune system1. Over the past few years, the prevalence of UC has significantly increased, particularly in the pediatric and elderly populations. It is widely recognized that UC results from the interaction and combined effects of various factors, including environmental factors, hereditary factors, microbes, and intestinal mucosal immunity2. Due to the complex pathogenesis, UC is often refractory and recurrent. Despite the advancements made in the management of UC inflammation and patient symptoms through the introduction of biologics, studies have shown that up to 30% of patients remain unresponsive to conservative drug treatments3,4. Therefore, early diagnosis and improved drug susceptibilities are critical clinical questions in the management of UC.
The polyamines, including putrescine, spermidine and spermine, are a class of positively charged small molecules distributing widely in various tissue5. They play a critical role in maintaining cellular viability by modulating various physiological processes such as maintaining DNA or RNA stability, promoting protein biosynthesis, and regulating cellular signal transduction6,7. Polyamine metabolism is a hub that interconnects the microbiota, diet, and immune system, and dysregulation of polyamine metabolism often leads to many diseases8. Clinical data suggest that polyamine levels are decreased in autoimmune diseases and inflammation9, and therapeutic interventions aimed at regulating polyamine metabolism have shown promise as a strategy for managing cancer, infection, autoimmune diseases, and inflammation10,11. Mechanistically, studies have demonstrated that spermine can inhibit the activation and proliferation of CD4+ T cells, as well as dose-dependently hinder the polarization of T effector cells12. Research conducted in vivo has shown that the absence of ODC1, which is the rate-limiting enzyme in the biosynthesis of polyamines, in group 3 innate lymphoid cells (ILC3s) can significantly decrease the severity of inflammatory colitis by reducing IL-22 production13. Furthermore, spermidine could improve intestinal barrier function and alleviate inflammation by alteration of microbiota composition and metabolite14. What’s more, as the main promoters of eIF5A hypusination, polyamines could rejuvenate memory B cell responses to by regulation of TFEB protein expression and autophagy15.
Given the essential and diverse functions of polyamines in immune regulation and disease pathogenesis, investigating polyamine homeostasis and developing novel therapies targeting polyamines could represent an intriguing and challenging area of research for UC. In the present study, we aimed to examine the relationship between polyamine-related genes (PRGs) and UC pathogenesis, and to investigate the potential correlations and underlying mechanisms between PRGs and the therapeutic effects of commonly used UC drugs. Our findings further enhanced the understanding of polyamine metabolism in UC and provided potential targets and implications for improving UC treatment.
Method
Data collections
To obtain mRNA-sequencing data and clinical data for this study, information on various datasets was collected from The Gene Expression Omnibus profile database (GEO) (https://www.ncbi.nlm.nih.gov/geo/) and described in detail in Table S1 (including the accession numbers). Data analysis was conducted using R software (version 4.2.1) (https://mirrors.tuna.tsinghua.edu.cn/CRAN/). We first performed a log transformation on the data, followed by normalization using the normalizeBetweenArrays function for GEO data sets. The R package “sva” was utilized to eliminate any batch effects present. Principal component analysis (PCA) was then performed to validate the results of batch correction and visualize the data.
Our study included a total of 137 polyamine-related genes (PRGs) that were obtained from multiple sources. These sources included the GeneCards human gene database (GeneCards—Human Genes | Gene Database | Gene Search), which was searched using the term “polyamine,” and genes with a relevance score > 1.5 were selected. The MSigDB database (GSEA (gsea-msigdb.org) GO: 1902267, R-HSA-351202) was also utilized to identify additional PRGs. Further details on the 137 PRGs used in this study can be found in Table S2.
Differential expression analysis
To identify differentially expressed genes (DEGs) in our study, the R package “limma” was utilized16. DEGs were determined between the UC-active group and both the healthy control group and the UC-inactive group using criteria of an adjusted p-value < 0.05 and |log2 fold change (FC)|> 1. The DEGs were then visualized using the R packages “pheatmap” and “ggplot2”.
Functional annotation and pathway enrichment analysis
We utilized the R packages “org.Hs.eg.db” and “clusterProfiler”17 to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.gsea-msigdb.org) analyses on the differentially expressed genes (DEGs) in our study. The results were then visualized using several R packages including “ggplot2”, “circlize”18, “enrichplot”, and “ComplexHeatmap”19.
Besides, differences between UC-active group and health control or UC-inactive group in metabolism pathway and immunologic function were analyzed by R package “GSEABase” and “GSVA”20. To identify significant pathways, we set the threshold for adjusted p-value to be less than 0.05. The results of the top 20 ranked pathways were then visualized using the R package “pheatmap”.
Developing a co-expression network and discovering gene modules
We utilized the R package “WGCNA” to build a weighted gene co-expression network21. To determine the appropriate soft threshold power (β), we performed scale-free topology model fitting and mean connectivity analysis. The adjacency matrix was then transformed into a topological overlap matrix (TOM) and subjected to average-linkage hierarchical clustering. Using the dynamic tree cut algorithm, we removed modules with fewer than 30 genes and clustered the remaining gene modules based on a cutoff value of 0.25. The genes within the same module had similar expression patterns and were associated with active UC. Finally, we identified hub genes in each module based on their gene significance (GS) greater than 0.5 and module membership (MM) greater than 0.8.
Discovery and validation of key polyamine-related genes associated with UC
We identified key PRGs associated with active UC by selecting genes that were present in all three categories: PRGs, DEGs, and hub genes in the most relevant module identified by WGCNA. To visualize the overlapping genes, we utilized the online tool “Calculate and draw custom Venn diagrams” (http://bioinformatics.psb.ugent.be/webtools/Venn/).
The diagnostic efficiency of the key PRGs was performed with receiver operating characteristic (ROC) analysis by R package “pROC”22.
Immune cell infiltration estimation
To investigate the immune cell composition in colon tissue of healthy control, inactive UC, and active UC groups, we utilized the CIBERSORT algorithm (https://cibersort.stanford.edu/)23. Only samples with P < 0.05 were selected for further analysis, and the relative abundance of 22 types of infiltrating immune cells was visualized using the R packages “corrplot” and “pheatmap”. In addition, the ssGSEA method was applied to calculate the immunity score and compare the clinical subgroups of immune cells. The results were presented using the R package “ggpubr”.
Single-cell analysis
Before the analysis, quality control of the single-cell sequencing data of immune cells from colonic lamina propria of active UC patients was performed using the R package “Seurat”24. We evaluated the sequencing depth (total number of UMIs, nCount), numbers of detected genes (nFeature) and the percentage of mitochondrial genes (percent.mt) in each sample. Samples with nFeature below 50 and percent.mt above 5% were excluded from subsequent analysis. After normalization of the filtered data, we identified 1500 highly variable genes (HVGs) by “vst” method. Based on these HVGs, PCA was applied to figure out marker genes in each principal component to reduce the dimension. In addition, genes that exhibited a |log2 FC|> 1 and adjusted p value < 0.05 were considered as the marker genes. Subsequently, t-distributed stochastic neighbor embedding (t-SNE) clustering was performed on the cell samples.
Cell culture and reagents
The HT29 cell lines were obtained from the Cell Bank of the Chinese Academy of Science (Shanghai, China) and cultured in 1640 medium (HyClone, USA) supplemented with 10% (v/v) fetal bovine serum (FBS, Gibco, USA), 0.1 mg/ml streptomycin (Biosharp, China) and 100U/ml penicillin (Biosharp, China) at 37 °C in a humidified incubator with 5% CO2. Lipopolysaccharides (LPS; HY-D1056), Azoxymethane (AOM; HY-111375) and Dextran sulfate sodium (DSS; HY-116282C) were purchased from MedChemExpress (MCE, USA). Spermine (S4264-1G), and spermidine (S2626-1G) were purchased from Sigma Aldrich (USA).
Animal experiments
DSS colitis mouse model: Male C57BL/6 mice, 8 weeks old, were used to establish an acute colitis model by providing ad libitum access to drinking water containing 3% DSS for 7 days. On the 8th day, mice were anesthetized with 2% pentobarbital and euthanized by cervical disloaction. Harvested colons for further analysis (Figure S1).
The AOM/DSS mouse model: Male C57BL/6 mice at 8 weeks old were used to establish the AOM/DSS model. The mice were first intraperitoneally injected with 10 mg/kg AOM (Sigma-Aldrich, MO, USA) and then provided with a normal diet and free access to water for 7 days. Afterward, they were given drinking water containing 1.5% DSS (Sigma-Aldrich, MO, USA) for 7 consecutive days, followed by 14 days of normal drinking water. This process was repeated three times to establish the AOM/DSS model. After fed a conventional diet for 10 weeks, the mice were euthanized by cervical dislocation, their colons are harvested for analysis (Figure S1).
All animal studies were approved by the Zhongnan Hospital Experiment Animal Welfare ethics committee affiliated to Wuhan University, and all methods were performed in accordance with the relevant guidelines and regulations.
RNA interference and gene transfection
The small interfering RNAs (siRNAs) targeting NNMT and scrambled control siRNA were provided by RiboBio (Guangzhou, China), and the sequences of NNMT siRNAs were as follows: siNNMT#1: GCAGAAAGCCAGATTCTTA; siNNMT#2: CGTCGTCACTGACTACTCA; siNNMT#3: CTTCCACCATGGCCAACAA. The NNMT siRNAs were transfected into HT29 cells using Lipofectamine 2000 transfection reagent (Invitrogen, Green Island, CA) according to the manufacturer’s protocol.
Quantitative real-time polymerase chain reaction (Q-PCR) analysis
Total RNA was isolated from cells or tissues using Trizol reagent (Invitrogen, USA) and reverse transcribed into cDNA using the reverse transcription PCR kit (TOYOBO, Japan). The expression levels of target genes were quantified by real-time PCR using UltraSYBR Mixture (CWbio, China) according to the manufacturer’s instructions. The 2−ΔΔCt method was used to calculate the relative change in mRNA levels, with GAPDH as the reference gene. The primer sequences used in this study were provided in Table S3.
Western blotting
The total proteins from cell samples were isolated using sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene fluoride membranes (ISEQ00010; Merck Millipore). Primary antibodies against ATP13A2 (1:1000, 5879 s; CST, USA), SMOX (1:1000, 15,052–1-AP; Proteintech, China), PAOX (1:1000, 18,972–1-AP; Proteintech, China), ODC (1:1000, 52238 s; CST, USA), IL6 (1:1000, 12912 s; CST, USA), IFN-γ (1:1000, 80385 s; CST, USA), TNF-α (1:1000, 11948 s; CST, USA), SOX2(1:1000, 11064-1-AP; Proteintech, China), Nanog (1:1000, 14295-1-AP; Proteintech, China), SOX4 (1:1000, ab70598; Abcam, USA), CD44 (1:1000, 15675-1-AP; Proteintech, China), CD133 (1:1000, 18470-1-AP; Proteintech, China), SAT1 (61586S, 1:1000; CST, USA), NNMT (1:1000, 15123-1-AP; Proteintech, China) and GAPDH (1:7500, 10494-1-AP; Proteintech, China) were separately incubated with the membranes overnight at 4 °C. Finally, the membranes were incubated with peroxidase-conjugated secondary antibodies against rabbit or mouse IgG (1:5000; Promoter, China) for 2 h at room temperature. The chemiluminescence imaging analysis was performed using the Genegenome XRQ system (Synoptics, version 4.0MP). The specific software and system versions utilized were as follows: Camera: Synoptics 4.0MP; GeneSys Version: 1.4.2.0; Database Version: 1.84. Blots were exposed for different durations to visualize specific target proteins.
Immunohistochemistry (IHC)
The tumor and intestinal tissues were fixed in 4% paraformaldehyde overnight, and then embedded in paraffin and sliced into sections. Deparaffinization was carried out using xylene, followed by rehydration with graded alcohol solutions. The sections were blocked with 3% Bull Serum Albumin (BSA), and then incubated with NNMT antibody (1:300, 15123-1-AP, Proteintech, China) at 4 °C overnight. Afterward, secondary antibodies were applied to the slides for 50 min at room temperature. The slides were then treated with diaminobenzidine (DAB) solution for color development, and counterstained with haematoxylin. Finally, the tissue slides were observed under a microscope (IX51, Olympus, Japan).
Statistical analysis
Statistical analysis was performed using R software (version 4.2.1) and Graphpad Prism 9 (Graphpad Software Inc., United States). The Unpaired Student’s t-test was applied for comparison of normally distributed variables between two groups, while one-way analysis of variance was used for three groups. A p-value < 0.05 was considered statistically significant.
Ethics approval and consent to participate
The study involving animal subjects was conducted under the approval of the Institutional Animal Care and Use Committee of Zhongnan Hospital of Wuhan University. The study involving human data was approved by the Ethical Committee of The Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology, with ethics exemption granted for the use of publicly-available human data in this research. All experiments were performed in accordance with relevant named guidelines and regulations, and this study is reported in accordance with the ARRIVE guidelines.
Result
The immune-infiltrating landscape in healthy control, UC_inactive and UC_active groups
Figure 1 illustrates the study process in the form of a flowchart.
Initially, we gathered data from three GEO cohorts (GSE48958, GSE53306, and GSE75214), which included bulk RNA sequencing information from healthy controls, inactive ulcerative colitis (UC), and active UC samples. To ensure accuracy and reliability, we merged the three GEO cohorts after correcting for batch effects, resulting in a comprehensive cohort of 31 healthy controls, 41 inactive UC, and 97 active UC samples (Fig. 2A,B). PCA indicated that the active UC samples were notably distinct from the other two groups (Fig. 2C). We then employed the CIBERSORT algorithm to compare the differences in immune cells among the three groups. Samples with p > 0.05 were considered reliable, and we obtained the relative percentages of 22 immune cells in 3 healthy controls, 10 inactive UC, and 68 active UC samples (Fig. 2D). Figure 2E depicted the correlations between the 22 immune cells, revealing that M2 macrophages were negatively correlated with activated mast cells (R = − 0.6), while follicular helper T cells were positively correlated with naïve B cells (R = 0.59) and resting dendritic cells (R = 0.65). Furthermore, CD8 + T cells were positively correlated with regulatory T cells (Tregs) (R = 0.61) and activated natural killer (NK) cells (R = 0.51). Finally, we compared the immune cell infiltrations and immune functions between the groups, as shown in Fig. 2F,G. As expected, the active UC group exhibited the highest level of immune cell infiltration and activation of several immune functions.
The immune-infiltrating landscape in healthy control, UC_inactive and UC_active groups. (A,B). PCA analysis showing the distribution of samples in GSE48958, GSE53306 and GSE75214 before (A) and after (B) batch effect correction. (C) PCA analysis showing the distribution of samples in healthy control, UC_inactive and UC_active groups. (D) The relative percent of 22 immune cells in each sample in the total set. (E) The correlation analysis between 22 immune cells based on the total set. Red and blue indicates a positive or negative correlation, respectively. The darker the color, the stronger the correlation. (F) Comparison of immune cell infiltrations in healthy control, UC_inactive and UC_active groups. (ns no significant, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001). (G) Immune function analysis between healthy control and UC_active groups (up), UC_inactive and UC_active groups (down). (R software (version 4.2.1) (https://mirrors.tuna.tsinghua.edu.cn/CRAN/)).
Functional enrichment analysis and polyamine metabolic reprogramming in healthy control, UC_inactive and UC_active groups
We employed a filtering approach, selecting differentially expressed genes (DEGs) with an adjusted p-value < 0.05 and |log2 FC|> 1 in both cohort A (consisting of healthy controls and active UC samples) (Fig. 3A,B) and cohort B (comprising inactive UC and active UC samples) (Figure S2). GO analysis revealed that immune-related pathways, such as cytokine-mediated signaling pathway, leukocyte migration, secretory granule membrane, secretory granule lumen, immune receptor activity, and cytokine activity, were enriched in the UC_active group (Fig. 3C,E). Similarly, KEGG analysis demonstrated enrichment of cytokine-cytokine receptor interaction, PI3K-Akt signaling pathway, and chemokine signaling pathway in the UC_active group in both cohort A (Fig. 3D) and cohort B (Fig. 3F).
Differential expression analysis and functional enrichment analysis between UC_active and healthy control group. (A,B). Volcano plots showing the DEGs between UC_active and healthy control (A) or UC_inactive group (B). (C,D) GO (C) and KEGG (D) analysis between healthy control and UC_active group. (E, F) GO (E) and KEGG (F) analysis between UC_inactive and UC_active group.
Apart from immune-related factors, nutrients and metabolites are also significant regulators of immune cell function25. When immune cells are activated, they undergo metabolic changes to meet the elevated energy requirements and sustain their immune functions26. The interplay between immune cells and metabolism orchestrates both innate and adaptive immunity26,27. Therefore, we analyzed the metabolic pathway profile between the UC_active and healthy control groups (Fig. 4A) or UC_inactive groups (Fig. 4B). We observed changes in numerous metabolic pathways in the UC_active group, including classical energy metabolism pathways, such as citric acid cycle, fatty acid degradation, and glycogen degradation. Interestingly, we also noticed that the polyamine biosynthesis pathway was active in the UC_active group. Recent studies have highlighted the significant role of polyamines in mammalian immune systems, particularly inflammation, although the underlying mechanism remains controversial 9. Hence, we selected polyamine metabolism for further investigation.
The difference on metabolic molecular signatures between UC_active and healthy control (A) or UC_inactive group (B). (R software (version 4.2.1) (https://mirrors.tuna.tsinghua.edu.cn/CRAN/)).
WGCNA to identify hub modules and hub genes associated with active UC
We performed WGCNA to identify hub modules and hub genes associated with active UC. After clustering analysis, no outlier sample was detected in the total set (Figure S3A,B). We selected 13 as the soft threshold power (β) in both cohort A and cohort B based on the scale-free topology model fit and mean connectivity (Figs. 5A and S4A). In cohort A, we identified 6 modules (Fig. 5B), and the red module had the strongest negative correlation with active UC (R = − 0.79, p < 0.001) (Fig. 5C). Therefore, we designated the red module as the hub module, and filtered out hub genes based on MM > 0.8 and GS > 0.5 (Fig. 5D, Table S4). In cohort B, we also identified 5 modules (Figure S4B), and the green module had the strongest positive correlation with active UC (R = 0.66, p < 0.001) (Figure S4C). Similarly, we defined the green module as the hub module in cohort B, and identified hub genes in the right upper quadrant of the module eigengene heatmap (Figure S4D, Table S5).
WGCNA to identify hub modules and hub genes based on healthy control and active UC. (A) Selection of the soft-thresholding powers. The left panel shows the relationship between soft-thresholding powers and scale free topology model fit, while the right shows the relationship between soft-thresholding powers and mean connectivity. (B) Module clustering dendrogram based on a dissimilarity measure (1-TOM). Different modules are colored differently. (C) Heatmap of the correlation between each module and trait. Red and blue indicates a positive or negative correlation, respectively. The darker the color, the stronger the correlation. (D) Scatter plot of the relationship between module membership (MM) and gene significance (GS) in red module. Genes in right upper quadrant are identified as hub genes in red module.
Identification and verification of the key PRGs associated with active UC
To identify key PRGs associated with active UC, we intersected DEGs, hub genes, and PRGs, resulting in 5 key genes in cohort A (Fig. 6A) and 2 key genes in cohort B (Figure S4E). We subsequently verified the expression of these key genes among healthy control, UC_inactive, and UC_active groups in the total set (Fig. 6B). The expressions of TGM2 and TRIM22 sequentially increased in healthy control, UC_inactive, and UC_active groups, while PPARG sequentially decreased. Additionally, the expressions of NNMT and PTGS2 in UC_active group were significantly higher than those in healthy control and UC_inactive groups. ROC curves showed that each key PRG had a significant clinical diagnostic value for active UC (Figs. 6C, S4F). We further confirmed these results with external validation sets GSE87473 and GSE107499, where differential expression analysis and ROC curves supported the key genes as biomarkers indicating active UC (Figure S5). We also investigated the relationships between key genes and immune infiltrating cells based on the total set. As shown in the heatmap (Fig. 6D), NNMT, PTGS2, TGM2, and TRIM22 had a positive association with most immune infiltrating cells, while PPARG had a negative association. Among the 5 key genes, NNMT and PTGS2 had much stronger correlation with immune cells, particularly type I Tregs, natural killer T cells, neutrophils, mast cells, and immature dendritic cells. These findings suggest that the identified 5 key genes could serve as potential biomarkers for active UC and are closely associated with immune infiltrating cells.
Identification and verification of the key PRGs in healthy control and active UC. (A) Venn diagram of the overlap among DEGs (between healthy control and UC_active group), hub genes in red module and PRGs. Genes in the center overlapping region are identified as key PRGs associated with active UC. (B) Differential expression analysis of key PRGs in healthy control, UC_inactive and UC_active groups. (ns no significant, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001). (C) ROC curves of each key PRG for diagnosis of active UC. (D) Heatmap of the correlation between each key PRG and immune cells. Red and blue indicates a positive or negative correlation, respectively. The darker the color, the stronger the correlation.
The role of key PRGs in therapeutic response of UC patients
Although significant advances have been made in the treatment of UC, there remains a group of patients who are unresponsive to existing therapies28. Therefore, we investigated the role of key genes in the therapeutic response of UC patients and evaluated their potential as biomarkers to improve drug sensitivity. Glucocorticoids (GCs) are essential drugs for combatting inflammatory diseases, and high doses of GCs are commonly administered to UC patients with severe symptoms or experiencing an acute relapse29,30. We compared the expressions of key PRGs in UC patients at different stages of treatment and found no significant difference in the intestinal mucosa of GC responders or non-responders before or after GC treatment (Figure S6A), suggesting that GCs may not affect these 5 key genes. With the advancements in therapeutic strategies for UC, we focused on emerging targeted biological agents31. Infliximab (IFX), the first TNF antagonist approved for treating UC, can induce and maintain remission and mucosal healing32,33. Golimumab, the third anti-TNF therapy for the treatment of moderate-to-severe UC, was also examined34. Vedolizumab (VDZ), a first-line biological targeting α4β7 integrin, was recommended for UC patients in the case of immunosuppressive or TNF antagonist therapy failure35. After treatment with IFX or VDZ, the expressions of all the key genes in responders were significantly different from those in non-responders, and the levels of expressions in responders after treatment were closer to those in healthy controls (Figs. 7, S6B). Specifically, compared with non-responders, NNMT, PTGS2, TGM2, and TRIM22 were remarkably decreased in the intestinal mucosa of responders receiving treatment, while PPARG was increased. However, despite being a TNF antagonist, golimumab had different effects on these 5 key genes compared to IFX. Differential expression analysis indicated that in golimumab-responders, the expressions of NNMT, TGM2, and PPARG were changed before and after golimumab treatment, while PTGS2 and TRIM22 did not differ across all groups (Figure S6C). In summary, the key genes remained unchanged with GCs treatment but showed variations with novel biological agents. This hints at a potential link between these genes and the effectiveness of biological agents, especially IFX and VDZ, in drug response. However, further experimental validation is needed to understand the underlying mechanisms.
Expression of key PRGs in colonic lamina propria immune cells of active UC by scRNA analysis
To further elucidate the associations between 5 key genes and active UC, we analyzed a single-cell RNA sequencing (scRNA-seq) database (GSE162335) consisting of information from colonic lamina propria immune cells from 11 active UC patients to identify the cell populations expressing key genes in active UC. Quality control indicated that all samples met the criteria, with more than 50 detected genes (nFeature) and less than 5% mitochondrial genes (Figure S7A). After preprocessing, a dataset comprising 18,179 high-quality cell samples was obtained, and a strong positive correlation was observed between nFeature and sequencing depth (total number of UMIs, nCount) (R = 0.86) (Figure S7B). Based on selected thresholds, 1500 variable genes were identified (Figure S7C), and PCA was utilized to decrease the dimensionality of the dataset by focusing on these variable genes (Figure S7D). After selecting the top 20 principal components to further reduce dimensionality, t-SNE method was used to cluster the cell samples, resulting in 20 subgroups (Figs. 8A, S8A). We calculated the contribution value of genes and defined marker genes for each subgroup, and the top 10 marker genes were visualized in the heatmap (Figure S8B).
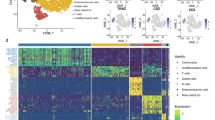
The cell annotation and the expression levels of key PRGs in colonic lamina propria immune cell types in active UC. (A,B) Visualisation of clustering (A) and annotation (B) in the tSNE plot. (C) The tSNE plots showing distribution of key PRGs in different immune cell types. (D,E) The violin plots (D) and bubble chart (E) describing expression levels and expression proportion of key PRGs in different immune cell types, respectively.
We then annotated and categorized cell samples into six cell types, including B cells, T cells, monocytes, common myeloid precursor cells (CMP), tissue stem cells, and macrophages (Fig. 8B). We found that T cells, B cells, and monocytes were the main immune infiltrating cells in the colonic lamina propria of active UC patients. We subsequently investigated the expression of key genes in different cell types (Fig. 8C). TRIM22 had the most extensive distribution, including B cells, T cells, monocytes, and tissue stem cells, while PPARG expression was low in all cell types and sporadically distributed in a subset of T cells (Fig. 8D,E). Significantly, PTGS2 was markedly enriched in CMP and monocytes, and NNMT and TGM2 were specifically expressed in tissue stem cells (Fig. 8D,E). These results highlight the close relationships between CMP, monocytes, tissue stem cells, and PRGs, and their crucial roles in colonic immunity, even though these cell types were present in small numbers.
Verification of diagnostic efficiency of the key PRGs in colitis-associated CRC (CAC)
Persistent inflammation is one of major causes of cancer development, which can lead to the formation of dysplastic precursor lesions in multiple regions of the colon via a phenomenon called field cancerization36. Given the increased risk of CAC development in patients with ulcerative colitis (UC), we evaluated the diagnostic potential of key genes for CAC.
We first merged two Gene Expression Omnibus (GEO) cohorts containing healthy controls and CAC patients after batch effect correction, resulting in a comprehensive cohort comprising 13 healthy controls and 26 CAC samples (Fig. 9A,B). PCA showed marked differences between CAC samples and healthy controls (Fig. 9C). Metabolism analysis indicated that both CAC and UC shared multiple changes in comparison to healthy controls, particularly in the active polyamine biosynthesis pathway (Fig. 9D). Notably, the expressions of NNMT, PTGS2, TGM2, and TRIM22 were significantly upregulated in CAC tumor tissue, while PPARG was downregulated (Fig. 9E), mirroring the results obtained for UC. ROC curves were used to evaluate the diagnostic significance of key genes in distinguishing between healthy controls and CAC (Fig. 9F). All five key genes showed AUC values above 0.70, with NNMT, PTGS2, and TGM2 exceeding 0.85, suggesting their potential role in CAC diagnosis.
Verification of diagnostic efficiency of the key PRGs in CAC. (A,B) PCA analysis showing the distribution of samples in GSE4183 and GSE37283 before (A) and after (B) batch effect correction. (C) PCA analysis showing the distribution of samples in healthy control and CAC groups. (D) The differences on metabolic reprogramming between healthy control and CAC groups. (R software (version 4.2.1) (https://mirrors.tuna.tsinghua.edu.cn/CRAN/)). (E) Differential expression analysis of key PRGs between healthy control and CAC groups. (ns no significant, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001). (F) ROC curves of each key PRG for CAC diagnosis.
Validation of the role of polyamines and NNMT in inflammation and CAC in vitro and vivo models
In order to explore the relationship between polyamines and intestinal inflammation, we conducted experiments in both in vitro and in vivo models. We detected changes in mRNA and protein levels of polyamine metabolism enzymes in HT29 cells treated with LPS. As shown in Fig. 10A,B, the expression of polyamine metabolism enzymes changed after LPS stimulation, suggesting that inflammation can lead to polyamine remodeling. Inflammatory conditions upregulated the polyamine transporter ATP13A2 and the polyamine synthesis enzymes ADM1 and ODC1, while downregulating to some extent the polyamine degradation enzymes SMOX and PAOX. These findings suggest that cells promote polyamine synthesis and inhibit polyamine degradation in order to increase intracellular polyamine levels and alleviate damage caused by inflammation. To further investigate the impact of polyamines on UC, we supplemented exogenous spermine and spermidine to HT29 cells after LPS stimulation and evaluated changes in inflammatory cytokines (Fig. 10C,D). The results demonstrated that polyamine supplementation significantly reduced the levels of inflammatory cytokines, suggesting that polyamine levels can directly inhibit inflammation. Furthermore, we investigated the expression of five key genes in the cell inflammation model (Fig. 10E), mouse colitis model (Fig. 10F), and mouse CAC model (Fig. 10G). Consistent with previous findings, NNMT, PTGS2, TGM2, and TRIM22 were upregulated to varying degrees in colitis and CAC models, while PPARγ was significantly downregulated. (As mice do not have an ortholog for the human TRIM22 gene, only the mRNA of the other four genes was detected in the mice).
Verification of the role of polyamines and key PRGs in inflammation or CAC. (ns no significant, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001). (A,B) The mRNA (A) and protein (B) levels of polyamine metabolic enzymes in HT29 treated with LPS. (C,D) The mRNA (C) and protein (D) levels of inflammatory cytokines in HT29 cells supplemented with exogenous spermine and spermidine following LPS stimulation. (E–G) The mRNA expression levels of key PRGs in HT29 treated with LPS (E), DSS-induced colitis model (F), and AOM/DSS-induced CAC model (G).
Based on our previous findings (Figs. 6A and S4E), NNMT and PTGS2 have been identified as effective diagnostic markers for both UC and control groups, with good diagnostic performance in both active and non-active UC. Given the well-established role of PTGS2 in inflammation, we focused our investigation on the impact of NNMT on UC and CAC. We verified the knockdown efficacy of three siRNAs targeting NNMT using qPCR and western blot (Fig. 11A,B), and identified siNNMT1#1 and #2 as the most effective for subsequent experiments. In LPS-treated HT29 cells, we observed that knockdown of NNMT significantly reduced cell viability and increased LPS-induced cell death (Fig. 11C). Furthermore, we examined the expression levels of inflammatory factors (Fig. 11D) and found that IL6, TGFβ, and TNFα were upregulated following NNMT knockdown, indicating a potential anti-inflammatory role for NNMT. We also detected the expression of NNMT1 in an AOM/DSS-induced CAC model and observed a significant upregulation of NNMT in tumor tissues (Fig. 11E). Given our previous observation of NNMT enrichment in tissue stem cells, we investigated the effect of NNMT on cell stemness. Our analysis of classical stem cell biomarkers (Fig. 11F) showed that knockdown of NNMT resulted in downregulation of multiple stem cell markers, including SOX4, SOX2, Nanog, and CD44, emphasizing the crucial role of NNMT in maintaining cell stemness.
Verification of the role of NNMT in inflammation and cell stemness. (ns no significant, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001) (A,B). The mRNA (A) and protein (B) levels of NNMT in HT29 transfected with siNNMT. (C) The effect of NNMT on cell viability was evaluated using the CCK8 assay in HT29 cells treated with LPS. (D) The mRNA levels of inflammatory cytokines in HT29 cells transfected with siNNMT following LPS stimulation. (E) The protein expression levels of NNMT in CAC tissue were detected with IHC. (F) The protein expression levels of stemness-related genes in HT29 cells transfected with siNNMT.
Discussion
UC is a chronic inflammatory bowel disease characterized by recurring periods of remission and relapse, and its etiology and pathogenesis are not yet fully understood37. The lack of understanding of the underlying mechanisms poses significant challenges for the treatment and prevention of UC. Recent studies suggest that polyamines may play a vital role in the immune system and can serve as prognostic indicators for autoimmune diseases8,9. In this study, we aimed to further investigate the diagnostic and therapeutic potential of polyamine-related genes in UC.
Previous studies have demonstrated the regulatory effect of polyamines on immunometabolism and inflammation. For instance, Wei et al. found that LACC1 regulates the interplay between proinflammatory NOS2 signaling and anti-inflammatory polyamine-mediated cytokine suppression in macrophages38. Additionally, Gobert et al. have shown that spermidine exerts a protective effect against colitis and colon carcinogenesis, suggesting its potential as an adjunctive treatment for colitis and chemopreventive agent for colon carcinogenesis39. To further investigate the roles of polyamines in UC, we identified five key PRGs and evaluated their potential as diagnostic markers for UC.
Nicotinamide N-methyltransferase (NNMT) is an intracellular enzyme that catalyzes the N-methylation of NAM using SAM, thereby regulating NAD-related signaling pathways, polyamine flux, and chromatin remodeling40. The role of NNMT in tumor progression is highly complex. Inhibition of NNMT upregulates the activity of ODC and SSAT, key enzymes in polyamine metabolism, increasing polyamine flux and energy consumption41. NNMT can also enhance SIRT1 activity by reducing its ubiquitination, promoting cell migration and invasion, and increasing cancer cell chemoresistance42. Additionally, NNMT acts as a regulator for the methyl donor to affect histone methylation, thereby impacting tumor epigenetics43. These pathways are interrelated; polyamine synthesis consumes acetyl-CoA, which functions as a substrate in histone acetylation, thus affecting the level of histone acetylation. Moreover, spermidine can induce the upregulation of Sirtuin 144. Therefore, the regulatory mechanisms of NNMT in tumors are highly intricate. Clinical and basic research has confirmed that high stromal NNMT expression predicts an unfavorable prognosis in colorectal cancer45, and is a reliable biomarker for targeted tumor therapies46. However, the exact role of NNMT in UC remains unclear. Our study suggests that NNMT can be used as a predictor of clinical response and disease progression in UC. Recently, research has shown that NNMT can maintain a high level of NAD-dependent proinflammatory signaling by removing excess inhibitory nicotinamide (Nam) in UC47, which corroborates our results. Furthermore, single-cell data analysis results indicate that NNMT is highly enriched in tissue stem cells, highlighting its role in self-renewal and differentiation. Several studies have reached conclusions consistent with ours. Pozzi V et al. showed that NNMT expression was associated with cancer stem cell enrichment48. Moreover, NNMT has been found to have an important role in regulating pluripotency of human embryonic stem cells (hESC)49. Given that strong stemness and pluripotency are typical characteristics of tumor cells, intervention of NNMT may block colitis-propelled carcinogenesis by inhibiting the stemness of cells.
Prostaglandin-endoperoxide synthase 2 (PTGS2), also known as cyclooxygenase 2 (COX-2), is a critical enzyme in the biosynthesis of prostaglandins (PGs) and is widely accepted as an indicator of inflammatory activity50. Our study showed that the expression of PTGS2 was significantly elevated in active UC patients compared to healthy controls and inactive UC patients, consistent with previous research that targeted delivery of celecoxib, a selective inhibitor of COX-2, effectively reduced the severity of UC51.
Tissue transglutaminase (TGM2) is a multifunctional enzyme that modifies proteins through an acyl transfer reaction, and previous studies have mainly focused on its role in cancer. For instance, Patrizia Malkomes et al. reported that TGM2 promotes the tumorigenicity of colon cancer cells by inactivating the tumor suppressor p5352. Besides, TGM2-mediated cross-linking of CXCL12 and KRT19 protein can suppress T cell–mediated immune attack and contribute to resistance to immunotherapy in colorectal cancer53. Although TGM2 has been extensively studied in colon cancer, its role in UC remains unclear. Our study found that the expression of TGM2 was positively correlated with the activity of UC, suggesting that TGM2 might be involved in the inflammatory process of UC.
Tripartite motif 22 (TRIM22), a member of the tripartite motif (TRIM) family, have emerged as an important player endowed with both antiviral effects and modulatory capacity of the innate immune response54. The specific role of TRIM22 in polyamine synthesis remains to be explored. Recent studies have shown that TRIM22 promotes the ubiquitination and degradation of NRF2, which regulates the expression of polyamine metabolizing enzyme genes, such as ODC and SAT55. Thus, we hypothesize that TRIM22 may be involved in polyamine synthesis through its ability to decrease NRF2 expression56. As an antiviral defense gene, the role of TRIM22 in UC was still uncertain. Our study broadened the function of TRIM22 by demonstrating that TRIM22 could be used as biomarker to predict UC and CAC. However, further research is needed to investigate whether TRIM22 is involved in the progression of UC through its regulation of polyamine metabolism.
Peroxisome proliferator-activated receptor gamma (PPARG) is a type of nuclear receptor that plays a crucial role in regulating gene expression related to energy metabolism, cellular development, and differentiation57. PPARG is generally downregulated during inflammation and oxidative stress and in many cancers58. Likewise, in our study, PPARG was significantly reduced in UC, and negatively correlated with most immune cells, which indicated that PPARG served as an immunosuppressive factor during the progression of UC. In context of biochemical mechanisms, it has been confirmed that PPARG, as a central coordinator of the mucus barrier, could protect intestinal epithelial cells from invasion in several ways59. Selective activation of PPARG using pioglitazone has been shown to improve intestinal epithelial barrier function, alleviate NLRP6 inflammasome dysfunction, and modulate the composition of gut microbiota60. Besides, microbiota-induced PPARG signaling maintained the homeostasis of gut microbiota by promoting the energy metabolism of colonic epithelial cells toward β-oxidation61. It was also worth noting that PPARG played an essential role in the regulation of SAT activity in HCT116 and Caco-2 cells62. Given that SAT1 is the key enzyme in polyamine catabolism, polyamine metabolism may also be an important pathway for PPARG in intestinal barrier protection.
As standard anti-inflammatory and immune modulator medications often fail to achieve satisfactory results in many UC patients, there is a pressing need to develop more specific predictive biomarkers of therapeutic response63. In our study, we observed a significant alteration in the expression levels of five key PRGs in the IFX or VDZ response group after treatment, while no significant change was observed in the IFX or VDZ non-response group. These findings indicate that monitoring the expression levels of these key PRGs may serve as an indicator of clinical response and contribute to the development of personalized treatment plans for UC. Previous clinical studies have revealed that under homeostatic conditions, the levels of polyamines are positively correlated with circulating TNF-α, which is the therapeutic target of IFX64. Studies have shown that there may be a link between polyamines and TNF-α, as TNF-α was found to increase putrescine levels in primary cultured fibroblast-like synoviocytes in the rheumatoid arthritis synovium65. Additionally, animal studies have indicated that the consumption of spermidine could inhibit TNF-α-induced inflammation through the activation of RIP1 deubiquitination66. These results suggest that polyamines may be effective in predicting therapeutic response and highlight their potential in regulating inflammation. Although the detection of polyamines is complex due to their instability, the measurement of PRG mRNA expression levels may provide a more feasible alternative. However, the current literature lacks consensus on the relationship between polyamines and α4β7 integrin, which is the therapeutic target of VDZ. Our results provide a novel perspective for the treatment of UC with VDZ, but f additional research is required to clarify the underlying mechanisms.
Certainly, our study has some limitations. Firstly, in the process of identifying key PRGs, we compared differentially expressed genes between uninflamed UC, inflamed UC, and healthy conditions. This method may select a large number of inflammation-related genes. While this approach aids in identifying PRGs associated with inflammation, we acknowledge that it may introduce certain methodological limitations. Secondly, we currently lack a comparison between UC and CAC to determine whether PRGs can serve as biomarkers to distinguish between UC and CAC. This issue requires further research. Additionally, much of our research is based on analyses of existing databases. Although we conducted some experimental validations, further experiments are necessary to clearly elucidate the regulatory relationships between PRGs and the onset, progression, and treatment of UC.
Conclusions
In summary, our study revealed the remodeling of polyamine metabolism in active UC and indicated alterations in polyamine levels directly affect inflammation levels, confirming the critical role of polyamines in UC. Through bioinformatics analysis, we identified five key PRGs and validated their impact on UC disease status, drug efficacy, and carcinogenesis. Overall, our findings provide valuable insights into the pathogenesis and potential therapeutic targets of UC, as well as the diagnostic and predictive biomarkers for UC and CAC.
Data availability
The original contributions presented in the study are included in the article/Supplementary Material. The RNA sequencing data used in the study can be found in the GEO database (https://www.ncbi.nlm.nih.gov/geo/), and PRGs were obtained from the GeneCards human gene database (GeneCards—Human Genes | Gene Database | Gene Search) and the MSigDB database (GSEA (gsea-msigdb.org) the accession numbers: GO:1902267, R-HSA-351202). The detail of GEO database and the GeneCards human gene database (including the accession numbers) was described in Tables S1 and S2. Any additional questions can be directed to the corresponding authors.
References
Buie, M. J. et al. Global hospitalization trends for Crohn’s disease and ulcerative colitis in the 21st century: A systematic review with temporal analyses. Clin. Gastroenterol. Hepatol. 21(9), 2211–2221 (2023).
Krugliak Cleveland, N., Torres, J. & Rubin, D. T. What does disease progression look like in ulcerative colitis, and how might it be prevented?. Gastroenterology 162, 1396–1408 (2022).
Abreu, M. T. et al. Efficacy and safety of maintenance ustekinumab for ulcerative colitis through 3 years: UNIFI long-term extension. J. Crohn’s Colitis 16, 1222–1234 (2022).
Spinelli, A. et al. ECCO guidelines on therapeutics in ulcerative colitis: Surgical treatment. J. Crohn’s Colitis 16, 179–189 (2022).
Soda, K. Overview of polyamines as nutrients for human healthy long life and effect of increased polyamine intake on DNA methylation. Cells 11, 164 (2022).
Terui, Y. et al. Polyamines protect nucleic acids against depurination. Int. J. Biochem. Cell Biol. 99, 147–153 (2018).
Bae, D.-H., Lane, D. J. R., Jansson, P. J. & Richardson, D. R. The old and new biochemistry of polyamines. Biochim. Biophys. Acta Gen. Subj. 1862, 2053–2068 (2018).
Holbert, C. E., Cullen, M. T., Casero, R. A. & Stewart, T. M. Polyamines in cancer: Integrating organismal metabolism and antitumour immunity. Nat. Rev. Cancer 22, 467–480 (2022).
Chia, T., Zolp, A. & Miska, J. Polyamine immunometabolism: Central regulators of inflammation, cancer and autoimmunity. Cells 11, 896 (2022).
Niu, F. et al. Arginase: An emerging and promising therapeutic target for cancer treatment. Biomed. Pharmacother. 149, 112840 (2022).
McCubbrey, A. L. et al. Polyamine import and accumulation causes immunomodulation in macrophages engulfing apoptotic cells. Cell Rep. 38, 110222 (2022).
Zheng, R., Kong, M., Wang, S., He, B. & Xie, X. Spermine alleviates experimental autoimmune encephalomyelitis via regulating T cell activation and differentiation. Int. Immunopharmacol. 107, 108702 (2022).
Peng, V. et al. Ornithine decarboxylase supports ILC3 responses in infectious and autoimmune colitis through positive regulation of IL-22 transcription. Proc. Natl. Acad. Sci. U. S. A. 119, e2214900119 (2022).
Ma, L. et al. Spermidine improves gut barrier integrity and gut microbiota function in diet-induced obese mice. Gut Microbes 12, 1832857 (2020).
Zhang, H. et al. Polyamines control eIF5A hypusination, TFEB translation, and autophagy to reverse B cell senescence. Mol. Cell 76, 110–125 (2019).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Yu, G., Wang, L.-G., Han, Y. & He, Q.-Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Gu, Z., Gu, L., Eils, R., Schlesner, M. & Brors, B. circlize Implements and enhances circular visualization in R. Bioinformatics 30, 2811–2812 (2014).
Gu, Z., Eils, R. & Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinform. 32, 2847–2849 (2016).
Hänzelmann, S., Castelo, R. & Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-Seq data. BMC Bioinform. 14, 7 (2013).
Langfelder, P. & Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 9, 559 (2008).
Robin, X. et al. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 12, 77 (2011).
Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457 (2015).
Gribov, A. et al. SEURAT: Visual analytics for the integrated analysis of microarray data. BMC Med. Genom. 3, 21 (2010).
Chapman, N. M. & Chi, H. Metabolic adaptation of lymphocytes in immunity and disease. Immunity 55, 14–30 (2022).
Chou, W.-C., Rampanelli, E., Li, X. & Ting, J.P.-Y. Impact of intracellular innate immune receptors on immunometabolism. Cell Mol. Immunol. 19, 337–351 (2022).
Chapman, N. M., Boothby, M. R. & Chi, H. Metabolic coordination of T cell quiescence and activation. Nat. Rev. Immunol. 20, 55–70 (2020).
Hanzel, J., Hulshoff, M. S., Grootjans, J. & D’Haens, G. Emerging therapies for ulcerative colitis. Expert Rev. Clin. Immunol. 18, 513–524 (2022).
Clarisse, D. & Beck, I. M. New tricks of our old friend the glucocorticoid receptor. Cell. Mol. Gastroenterol. Hepatol. 11, 1541–1542 (2021).
Muzzi, C. et al. The glucocorticoid receptor in intestinal epithelial cells alleviates colitis and associated colorectal cancer in mice. Cell. Mol. Gastroenterol. Hepatol. 11, 1505–1518 (2021).
Kucharzik, T., Koletzko, S., Kannengiesser, K. & Dignass, A. Ulcerative colitis-diagnostic and therapeutic algorithms. Dtsch. Arztebl. Int. 117, 564–574 (2020).
Luo, H. et al. Emerging pharmacotherapy for inflammatory bowel diseases. Pharmacol. Res. 178, 106146 (2022).
Schmitt, H., Neurath, M. F. & Atreya, R. Role of the IL23/IL17 pathway in Crohn’s disease. Front. Immunol. 12, 622934 (2021).
Lasa, J. S., Olivera, P. A., Danese, S. & Peyrin-Biroulet, L. Efficacy and safety of biologics and small molecule drugs for patients with moderate-to-severe ulcerative colitis: A systematic review and network meta-analysis. Lancet Gastroenterol. Hepatol. 7, 161–170 (2022).
Dulai, P. S. et al. Decision support tool identifies ulcerative colitis patients most likely to achieve remission with vedolizumab vs adalimumab. Inflamm. Bowel Dis. 28, 1555–1564 (2022).
Shah, S. C. & Itzkowitz, S. H. Colorectal cancer in inflammatory bowel disease: Mechanisms and management. Gastroenterology 162, 715–730 (2022).
Nakase, H., Sato, N., Mizuno, N. & Ikawa, Y. The influence of cytokines on the complex pathology of ulcerative colitis. Autoimmun. Rev. 21, 103017 (2022).
Wei, Z., Oh, J., Flavell, R. A. & Crawford, J. M. LACC1 bridges NOS2 and polyamine metabolism in inflammatory macrophages. Nature 609, 348–353 (2022).
Gobert, A. P. et al. Protective role of spermidine in colitis and colon carcinogenesis. Gastroenterology 162, 813–827 (2022).
Wang, W., Yang, C., Wang, T. & Deng, H. Complex roles of nicotinamide N-methyltransferase in cancer progression. Cell Death Dis. 13, 267 (2022).
Kraus, D. et al. Nicotinamide N-methyltransferase knockdown protects against diet-induced obesity. Nature 508, 258–262 (2014).
Wang, Y. et al. Nicotinamide N-methyltransferase enhances chemoresistance in breast cancer through SIRT1 protein stabilization. Breast Cancer Res. 21, 64 (2019).
Ulanovskaya, O. A., Zuhl, A. M. & Cravatt, B. F. NNMT promotes epigenetic remodeling in cancer by creating a metabolic methylation sink. Nat. Chem. Biol. 9, 300–306 (2013).
Liu, X. et al. Spermidine inhibits vascular calcification in chronic kidney disease through modulation of SIRT1 signaling pathway. Aging Cell. 20, e13377 (2021).
Song, M. et al. High stromal nicotinamide N-methyltransferase (NNMT) indicates poor prognosis in colorectal cancer. Cancer Med. 9, 2030–2038 (2020).
Changqing, W. et al. NNMT‐DNMT1 axis is essential for maintaining cancer cell sensitivity to oxidative phosphorylation inhibition. Adv. Sci. https://doi.org/10.1002/advs.202202642 (2022).
Wnorowski, A., Wnorowska, S., Kurzepa, J. & Parada-Turska, J. Alterations in kynurenine and NAD+ salvage pathways during the successful treatment of inflammatory bowel disease suggest HCAR3 and NNMT as potential drug targets. IJMS 22, 13497 (2021).
Pozzi, V. et al. Cancer stem cell enrichment is associated with enhancement of nicotinamide N-methyltransferase expression. IUBMB Life 72, 1415–1425 (2020).
Novak Kujundžić, R. et al. Nicotinamide N-methyltransferase in acquisition of stem cell properties and therapy resistance in cancer. IJMS 22, 5681 (2021).
Hirano, T. et al. Immunological mechanisms in inflammation-associated colon carcinogenesis. IJMS 21, 3062 (2020).
Mishra, R. K. et al. Lipid-based nanocarrier-mediated targeted delivery of celecoxib attenuate severity of ulcerative colitis. Mater. Sci. Eng. C 116, 111103 (2020).
Malkomes, P. et al. Transglutaminase 2 promotes tumorigenicity of colon cancer cells by inactivation of the tumor suppressor p53. Oncogene 40, 4352–4367 (2021).
Wang, Z. et al. Carcinomas assemble a filamentous CXCL12–keratin-19 coating that suppresses T cell–mediated immune attack. Proc. Natl. Acad. Sci. U. S. A. 119, e2119463119 (2022).
Vicenzi, E. & Poli, G. The interferon-stimulated gene TRIM22: A double-edged sword in HIV-1 infection. Cytokine Growth Factor Rev. 40, 40–47 (2018).
Liu, W. et al. TRIM22 inhibits osteosarcoma progression through destabilizing NRF2 and thus activation of ROS/AMPK/mTOR/autophagy signaling. Redox Biol. 53, 102344 (2022).
Smirnova, O. A. et al. Chemically induced oxidative stress increases polyamine levels by activating the transcription of ornithine decarboxylase and spermidine/spermine-N1-acetyltransferase in human hepatoma HUH7 cells. Biochimie 94, 1876–1883 (2012).
Hayashi, Y. & Nakase, H. The molecular mechanisms of intestinal inflammation and fibrosis in Crohn’s disease. Front. Physiol. 13, 845078 (2022).
Vallée, A. & Lecarpentier, Y. Crosstalk between peroxisome proliferator-activated receptor gamma and the canonical WNT/β-catenin pathway in chronic inflammation and oxidative stress during carcinogenesis. Front. Immunol. 9, 745 (2018).
Fang, J., Wang, H., Xue, Z., Cheng, Y. & Zhang, X. PPARγ: The central mucus barrier coordinator in ulcerative colitis. Inflam. Bowel Dise. 27, 732–741 (2021).
Li, J.-M. et al. Dietary fructose-induced gut dysbiosis promotes mouse hippocampal neuroinflammation: A benefit of short-chain fatty acids. Microbiome 7, 98 (2019).
Byndloss, M. X. et al. Microbiota-activated PPAR-γ signaling inhibits dysbiotic Enterobacteriaceae expansion. Science 357, 570–575 (2017).
Ulrich, S. et al. Peroxisome proliferator-activated receptor γ as a molecular target of resveratrol-induced modulation of polyamine metabolism. Cancer Res. 66, 7348–7354 (2006).
Elhag, D. A. et al. Inflammatory bowel disease treatments and predictive biomarkers of therapeutic response. IJMS 23, 6966 (2022).
Liu, M. et al. Glutathione, polyamine, and lysophosphatidylcholine synthesis pathways are associated with circulating pro-inflammatory cytokines. Metabolomics 18, 76 (2022).
Won, W. et al. Inhibiting peripheral and central MAO-B ameliorates joint inflammation and cognitive impairment in rheumatoid arthritis. Exp. Mol. Med. 54, 1188–1200 (2022).
Chen, Z. et al. Spermidine activates RIP1 deubiquitination to inhibit TNF-α-induced NF-κB/p65 signaling pathway in osteoarthritis. Cell Death Dis. 11, 503 (2020).
Funding
This research was funded by the Hubei Provincial Natural Science and Technology Fund (No: 2023AFB458), Project of Wuhan Municipal Health Commission (NO.WX23Q10) and Wuhan Science and Technology Bureau project (No: 2023020201010186).
Author information
Authors and Affiliations
Contributions
Conceptualization, W.W., Y.L., and H.Z.; methodology, W.W., and Y.L.; software, W.W., Y.L., and M.Z.; validation, W.W., and J.G.; formal analysis, W.W.; investigation, M.Z.; data curation, W.W., and J.G.; writing—original draft preparation, W.W.; writing—review and editing, W.W. and H.Z.; visualization, W.W.; supervision, H.Z. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wei, W., Lu, Y., Zhang, M. et al. Identifying polyamine related biomarkers in diagnosis and treatment of ulcerative colitis by integrating bulk and single-cell sequencing data. Sci Rep 14, 18094 (2024). https://doi.org/10.1038/s41598-024-69322-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-69322-6
- Springer Nature Limited