Abstract
Formalin-fixed paraffin-embedded (FFPE) tissue represents a valuable source for translational cancer research. However, the widespread application of various downstream methods remains challenging. Here, we aimed to assess the feasibility of a genomic and gene expression analysis workflow using FFPE breast cancer (BC) tissue. We conducted a systematic literature review for the assessment of concordance between FFPE and fresh-frozen matched tissue samples derived from patients with BC for DNA and RNA downstream applications. The analytical performance of three different nucleic acid extraction kits on FFPE BC clinical samples was compared. We also applied a newly developed targeted DNA Next-Generation Sequencing (NGS) 370-gene panel and the nCounter BC360® platform on simultaneously extracted DNA and RNA, respectively, using FFPE tissue from a phase II clinical trial. Of the 3701 initial search results, 40 articles were included in the systematic review. High degree of concordance was observed in various downstream application platforms. Moreover, the performance of simultaneous DNA/RNA extraction kit was demonstrated with targeted DNA NGS and gene expression profiling. Exclusion of variants below 5% variant allele frequency was essential to overcome FFPE-induced artefacts. Targeted genomic analyses were feasible in simultaneously extracted DNA/RNA from FFPE material, providing insights for their implementation in clinical trials/cohorts.
Similar content being viewed by others
Introduction
Breast cancer (BC) represents a heterogeneous disease both clinically and biologically. Gene sequencing and gene expression profiling have emerged as important pillars in diagnostics and disease classification, biomarker discovery and treatment strategies in early and metastatic BC1. For such studies, archival formalin-fixed paraffin-embedded (FFPE) remains one of the main valuable patient tissue material sources for translational research and/or clinical applications and often is the only available patient tissue source. However, the use of nucleic acids derived from FFPE material for downstream applications has been challenging mainly due to pre-analytical setbacks, including tissue handling, extraction protocol, storage conditions, age, biospecimen size, formalin composition, delay of fixation and time in fixative2.
Given that the isolated nucleic acids are prone to degradation3,4, proper preservation of the biospecimen is of utmost importance to prevent risk of contamination, cell degeneration and nucleic acid degradation5. The most cost-efficient method for prolonged storage of clinical samples for a long time period, at ambient temperature and nonsterile conditions remains the use of formalin6. During fixation, a cross-linking occurs between formaldehyde and proteins, where formaldehyde triggers the formation of stable methylene bridges, thus making the tissue harsher7. Considering that nucleic acids are generally degraded due to the cross-linking and fixation process, FFPE tissue could therefore be less optimal for molecular downstream applications compared to fresh-frozen (FF) tissue8,9,10. Another factor that can affect the yield and the quality of the nucleic acids isolated from FFPE material is the extraction method. Several commercially available extraction kits offer robust performance but lack analytical validity and optimisation in different disease settings11. Thus, improving the yield and quality of nucleic acids extracted from FFPE becomes a pertinent issue to facilitate costly downstream analyses.
In this study, we aimed (i) to evaluate the degree of concordance between FFPE and FF material in matched BC tissue samples for certain RNA and DNA applications in published literature, (ii) to demonstrate the feasibility of simultaneous DNA and RNA extraction method on FFPE material using commercially available kits and (iii) to explore its performance on the hybridization-based nCounter® platform for gene expression analysis and on an in-house designed targeted NGS panel for genomic analysis using archival material from a randomized phase II early BC clinical trial.
Material and methods
Systematic literature search, inclusion criteria, data extraction and analysis
In order to identify studies evaluating the degree of concordance of the performance of downstream applications on the extracted nucleic acids between FF and FFPE matched BC tissue material, we conducted a systematic literature review in accordance with the PRISMA 2020 statement12. The systematic electronic search was performed in the following databases: Medline (Ovid), Embase, Web of Science (Clarivate), and PubMed Central. The original search was performed by two librarians at the Karolinska Institutet University Library on February 5th 2021 and the search was updated on February 2nd 2023 using the methods described by Bramer and Bain13. The search strategy was developed in Medline (Ovid). For each search concept, Medical Subject Headings (MeSH-terms) and free-text terms were identified. The search was then translated into the other databases. Language was restricted to English and databases were searched from inception. The strategies were peer-reviewed by another librarian prior to execution. De-duplication was performed as previously described14. The full search strategies for all databases are available as Supplementary Material S1. Three additional sources were used to ensure that all relevant articles were included: (i) the references of selected review articles on the topic were reviewed; (ii) secondary referencing by manually reviewing reference lists of potentially eligible articles; (iii) the Biospecimen Research Database (BRD) [http://biospecimens.cancer.gov/brd. Bethesda (MD), National Cancer Institute]. Studies were included in the systematic review only if they fulfilled the following criteria: studies that used nucleic acids (DNA and/or RNA) extracted from FF and FFPE matched tissue samples, derived from patients with BC, and applied certain technology platforms or methods i.e. microarray-based/DASL (cDNA-mediated annealing, extension, selection and ligation assay), multiplex hybridization nCounter®, RNA-sequencing technology platforms for RNA and Sanger and Next-Generation Sequencing for DNA. Case reports, reviews, prior systematic reviews, animal studies, conference material, editorials, letters and notes were excluded. Data extraction was performed using a predefined form for each study including the following variables: first author’s name, name of the journal, date of publication, type of nucleic acid (DNA/RNA); number of matched FF-FFPE tissue samples derived from patients with BC, degree of concordance reported as coefficient of determination (\(R^{2}\)), Spearman and/or Pearson correlation coefficients or Lin’s concordance correlation coefficient, or descriptively as reported by the authors when the same technology platform and analysis was performed using nucleic acids extracted from matched FF-FFPE BC tissue material. The title and abstract screening, full-text screening, and data extraction were performed by two investigators (D.S., I.Z.) and any discrepancies were resolved by a third investigator (A.M.).
Patient material
Archival surgical FFPE tissue material was used from thirty patients enrolled in the Scandinavian Breast Group (SBG) 2004-1 multicenter randomized phase II clinical trial. The trial enrolled a total of 124 patients with high-risk early BC and evaluated the feasibility of three adjuvant chemotherapy regimens: dose-dense epirubicin/cyclophosphamide (EC) followed by dose-dense docetaxel; the same regimen with additional tailored dosing according to hematologic toxicity during the previous treatment cycle; or concomitant docetaxel, doxorubicin and cyclophosphamide. The study design, feasibility and short-term toxicity15, long-term efficacy16 and immunogenomic analyses based on tissue material from the study17 have been previously reported. This trial was initiated in 2004, when trial registration was not compulsory. It is the feasibility study of the randomized phase III trial (PANTHER18, EudraCT number 2007-002061-12 and Clinicaltrials.gov accession number NCT00798070). All correlative analyses for this clinical trial have been approved by the Ethics Committee at Karolinska Institutet (Dnr 2017/345-32 and Dnr 2018/1084-32). In addition, anonymized archival surgical FFPE tissue material from patients with primary breast cancer (dated from 1992 to 2015) was used to assess the analytical performance of different commercially available DNA and RNA extraction kits, as described hereunder.
FFPE tissue preparation, block annotation and sectioning
Surgical FFPE BC tissue blocks were preserved at 4 \(^\circ\)C, after tissue fixation with 10% neutral-buffered formalin and paraffin embedment. Hematoxylin and eosin-stained sections 4 \(\upmu\)m in thickness from each FFPE BC tissue block were obtained and tumour-rich areas were annotated by a certified pathologist (J.H.). The tumour-rich blocks were subsequently sectioned (thickness: 10 \(\upmu\)m), using the Epredia™ HM 355S Automatic Microtome (Thermo Fisher Scientific, USA) and stored at −20 \(^\circ\)C until nucleic acid isolation.
Nucleic acid extraction from FFPE BC tissue samples
Tumour-rich surgical FFPE BC tissue blocks were cut into sections, each of 10 \(\upmu\)m in thickness. Two consecutively cut sections were used as starting material for each extraction protocol kit. The sections were then deparaffinized using xylene prior the purification of DNA and/or RNA. DNA extraction was performed using the QIAamp DNA FFPE Tissue kit (Cat No 56404, QIAGEN, Germany) and the AllPrep DNA/RNA FFPE kit (Cat No 80234, QIAGEN, Germany) according to the manufacturer’s instructions. However, DNA was eluted in Buffer EB (Cat No 19086, QIAGEN, Germany) instead of EDTA-containing Buffer ATE in order to prevent any enzymatic inhibition. Total RNA was extracted using the RNeasy FFPE kit (Cat No 73504, QIAGEN, Germany) and the AllPrep DNA/RNA FFPE kit, according to the manufacturer’s instructions (by also including small RNAs) and eluted in RNase-free water. To minimize the risk of any potential contamination in the RNA purification, DNase I digestion step was performed using the RNase-free DNase set (Cat No 79254, QIAGEN, Germany). For DNA purification, RNase A (100 mg/mL) (Cat No 19101, QIAGEN, Germany) step was performed accordingly. All nucleic acids were collected in Eppendorf® Forensic DNA Grade Safe-Lock 1.5 mL microcentrifuge tubes (Eppendorf SE, Germany), upon double elution at 20,000g, and stored at − 80 \(^\circ\)C. For every patient with BC, tumour DNA and RNA were extracted from two tumour-rich surgical FFPE tissue sections using only the AllPrep DNA/RNA FFPE kit. Tumour DNA and RNA were extracted from thirty patients enrolled in the SBG 2004-1 study using only the AllPrep DNA/RNA FFPE kit, while matched germline DNA was extracted from frozen whole peripheral blood, previously stored in EDTA-tubes at − 80 \(^\circ\)C, using FlexiGene DNA kit (Cat No 51206, QIAGEN, Germany) according to the manufacturer’s instructions.
Quality control and nucleic acid yield estimation
The initial quality control (QC) included: (i) the Nanodrop™ ND-1000 (Saveen Werner, Sweden), ultraviolet spectrophotometer to assess the purity19 of DNA and RNA samples based on the absorbance maximum at 260 nm (\(A_{260}\)) for nucleic acids, at 280 nm (\(A_{280}\)) for proteins, at 230 nm (\(A_{230}\)) for organic and salt isolation compounds, (ii) the Qubit® 3.0 Fluorometer (Thermo Fisher Scientific, USA) as well as the Qubit™ dsDNA BR Assay Kit (Cat No Q32850, Invitrogen, USA) and Qubit™ RNA BR Assay Kit (Cat No Q10210) to quantify the DNA and the RNA samples, respectively, and (iii) the automated electrophoresis 2200 Tapestation® System (Agilent Technologies, Santa Clara, CA, USA) and the Genomic DNA (Cat No 5067–5365) and RNA (Cat No 5067–5576) ScreenTapes to estimate the DNA Integrity Number (DIN) for DNA samples and RNA Integrity Number equivalent (RIN\(^{e}\)) for RNA samples, respectively. An additional quality metric DV\(_{200}\) for the RNA samples was generated manually, by using the raw data as presented in the region table in TapeStation Analysis Software, as follows: DV\(_{200}\) = (Number of RNA fragments with sizes between 200 nt and 10,000 nt / Total number of RNA fragments) * 100. The yield comparison of RNA extraction kits (RNeasy and AllPrep) was done after performing paired t-test for the RNA samples extracted from 7 FFPE blocks. Similarly, the yield comparison of DNA extraction kits (AllPrep and QIAamp) was done after performing paired t-test for the DNA samples extracted from 5 FFPE blocks.
Targeted DNA sequencing and bioinformatics analysis
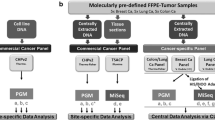
Tumour DNA and matched germline DNA samples derived from 30 patients with breast cancer enrolled in SBG2004-1 clinical trial were sequenced using a newly designed targeted DNA panel. This custom-developed DNA panel consisting of 370 genes (also referred to as GMCK Solid Cancer Panel) was used within the NovaSeq™ 6000 system (Illumina Inc.) with a 1000x average coverage. This targeted DNA GMCK Solid Cancer Panel (v1.0), 2.4 Mb in total size, was designed using the reference genome hg19, providing the rationale for identification of somatic short variants (Single-nucleotide Polymorphisms and Short Insertions and Deletions, SNVs/INDELs), Copy-number Alterations (CNAs), fusion events, Microsatellite Instability (MSI) and estimation of the Tumour Mutational Burden (TMB). Detailed description of the GMCK Solid Cancer Panel v1.0 panel is provided as Supplementary Material S1.
The bioinformatics analysis can be summarized as follows. Pre-processing following BALSAMIC workflow v9.0.120 was used to analyze each of the FASTQ files. Firstly, a quality control of FASTQ files using FastQC v0.11.9 was performed21. Adapter sequences and low quality bases were trimmed using fastp v0.23.222. Trimmed reads were mapped to the reference genome hg19 using BWA MEM v0.7.1723. The resulted SAM files were converted to BAM files and sorted using samtools v1.15.124,25. Duplicated reads were marked using Picard tools MarkDuplicates v2.27.126 and promptly quality controlled using CollectHsMetrics, CollectInsertSizeMetrics, and CollectAligntmentSummaryMetrics functionalities. Results of the quality controlled steps were summarized by MultiQC v1.1227. For each sample, somatic mutations were called using VarDict v2019.06.0428 in tumour-normal mode and annotated using Ensembl VEP v104.329. Apart from VarDict’s internal filters to report the variants, the called variants were also further post-filtered based on depth, frequency and quality, according to two filters followed in Blue Collar Bioinformatics (bcbio-nextgen) https://zenodo.org/records/5781867. Specifically, the first filter looks at regions with low depth for allele frequency (AF * DP < 6), and within these calls, it filters if a call has low mapping quality and multiple mismatches in a read ((AF * DP < 6) && ((MQ < 55.0 && NM > 1.0) || (MQ < 60.0 && NM > 2.0) || (DP < 10) || (QUAL < 45))). The second one filters in low allele frequency regions with poor quality if all of these are true: ((AF < 0.2) & (QUAL < 55) & (SSF > 0.06)). Only those variants that fulfilled the filtering criteria and scored as PASS in the VCF file were reported. Due to previously reported FFPE representative, artefactual C-T conversions that result from cytosine deamination during formalin fixation30,31 an additional filtering criterion was applied to exclude variants below 5% variant allele frequency32. Different post-filtering strategies could also be applied, e.g., different quality criteria or utilizing population databases, like the gnomAD database33 and the COSMIC somatic mutation database34, but this goes beyond the scope of the current feasibility study.
Mutational spectrums were identified using the R package MutationalPatterns v.1.2.135, and de-novo signatures were extracted based on the non-negative matrix factorization (NMF) algorithm.
nCounter® Breast Cancer 360™ Panel
RNA extracted from two patients with BC, previously enrolled in the SBG2004-1 clinical trial was used for expression profiling 776 gene targets using the nCounter® Breast Cancer 360™ gene panel (Nanostring Technologies, Seattle, USA) according to manufacturer’s instructions. Both the quality control and data pre-processing were performed in the nSolver Analysis Software version 4.0 (Nanostring Technologies, Seattle, USA). The raw data of the assay were assessed using quality assurance metrics to measure imaging quality, oversaturation, and overall signal-to-noise ratio. Background thresholding was performed with the default cut-off value of 20. The background-corrected data were then normalized using the geometric mean for two normalization factors, i.e., the 6 positive controls and the 18 housekeeping genes included in the assay.
Results
Literature search and study characteristics
A total of 3692 records were identified from the four databases (Medline, Embase, Web of Science and PubMed Central) and 9 records were identified via other methods (5 from Biospecimen Research Database and 4 from citation searching). Upon de-duplication, 2204 records identified from Databases, 131 were retrieved for full-text review and 40 articles were included in the review. The flowchart of study selection is presented in Fig. 1. Among the selected studies using matched FF-FFPE BC tissue samples, 31 articles evaluated only the performance of RNA, 8 articles only the performance of DNA and one article reported the performance of both RNA and DNA. All studies are presented in Tables 1 and 2, respectively.
Among the 32 studies that explored the performance of RNA extracted from matched FF-FFPE BC tissue material (Table 1) the degree of concordance was not reported in only 5 studies. Five studies used cDNA-mediated Annealing, extension, Selection and Ligation assay (DASL) while 9 studies used other microarray-based applications and reported varying concordance. Hybridization-based assays were used in 7 studies (6 studies used Nanostring nCounter® and 1 study used QuantityGene Plex assay) that reported high and excellent correlations on different probes (ranging from r = 0.66 and also exceeding r > 0.9; R2 = 0.89–0.96; CCC = 0.98). RNA-seq was used in 15 studies that reported high correlations on different levels (ranging from r = 0.589 and exceeding r > 0.9; R2 > 0.8; CCC = 0.63–0.96).
Among the nine studies that explored the performance of DNA extracted from matched FF-FFPE BC tissue material (Table 2), the degree of concordance was not reported in three studies. DNA sequencing with chain-terminating inhibitors (Sanger) was used in two studies, however the degree of concordance was not reported in either study. Next-Generation sequencing was used in five studies that reported varying correlations on different genome levels ranging from poor concordance in low-sequenced regions to high and excellent concordance in more reliable genome regions.
Analytical performance of different DNA/RNA extraction protocols in archival FFPE patient material
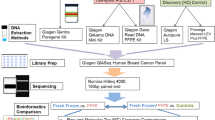
Based on the aforementioned results, we aimed to evaluate and compare mainly the nucleic acid yield using three different commonly used and commercially available kits for DNA only, RNA only and simultaneous DNA/RNA extraction from archival surgical FFPE tissue material derived from patients with early BC. The overall experimental workflow is depicted in Fig. 2 and the selection of the kits was based on the studies previously reporting high degree of concordance. RNA was extracted from 7 surgical FFPE tissue samples using consecutive sections for both RNeasy FFPE (RNeasy) and AllPrep DNA/RNA FFPE (AllPrep) kits. Quantification using the Nanodrop spectrophotometer revealed that the AllPrep extraction kit yielded slightly higher RNA compared to RNeasy (mean: 245.4 ng/\(\upmu\)L vs 209.8 ng/\(\upmu\)L, respectively) (paired t-test p = 0.553), but similar RNA yield based on Qubit fluorometer (mean: 175.4 ng/\(\upmu\)L vs 182.6 ng/\(\upmu\)L, respectively) (paired t-test p = 0.873). DNA was extracted from 5 surgical FFPE tissue samples using consecutive sections for both QIAamp DNA FFPE Tissue (QIAamp) and AllPrep extraction kits. Yield estimation based on Nanodrop spectrophotometer showed that the AllPrep extraction kit yielded similar DNA compared to QIAamp (mean: 130.6 ng/\(\upmu\)L vs 125.6 ng/\(\upmu\)L, respectively) (paired t-test p = 0.851). Similar yield between the AllPrep and QIAamp was also estimated based on Qubit fluorometer (mean: 61.8 ng/\(\upmu\)L vs 49.6 ng/\(\upmu\)L, respectively) (paired t-test p = 0.425). More information regarding the age, series of consecutive sections, material type and all initial QC metrics are available as Supplementary Material S1.
Experimental workflow including (a) the evaluation of three commercially available kits for the extraction of nucleic acids from FFPE BC tissue material sole DNA (QIAamp), sole RNA (RNeasy), and simultaneously DNA and RNA (AllPrep). The AllPrep and the RNeasy were used for nucleic acid extraction of seven (n = 7) archival FFPE BC tissue blocks, while the QIAamp was used only for DNA extraction from five (n = 5) of these seven FFPE blocks. The extracted nucleic acids underwent an Initial Quality Control evaluation using Nanodrop spectrophotometer, Qubit fluorometer and 2200 Tapestation automated electrophoresis system. (b) Tumour DNA extracted from thirty (n = 30) tumour-rich surgical FFPE BC tissue blocks derived from the SBG 2004-1 phase II clinical trial using the AllPrep, followed by targeted DNA GMCK panel, while matched germline DNA extracted from whole peripheral blood using FlexiGene DNA kit was used as control. Tumour RNA samples (n = 2) were analyzed using the Human nCounter Breast Cancer 360 gene panel and the nSolver Analysis Software version 4.0.
Performance of the simultaneous DNA/RNA extraction protocol and targeted DNA sequencing in archival FFPE clinical samples
Considering the similar performance among the extraction kits, solely based on the initial QC metrics, we aimed to demonstrate the feasibility of downstream applications for the simultaneously extracted DNA/RNA using AllPrep, comparable to the findings of the systematic literature review. We thus used archival surgical FFPE tissue and matched whole peripheral blood from thirty patients previously enrolled in the SBG2004-1 study. After using AllPrep kit and two FFPE sections of 10 \(\upmu\)m in thickness as starting material for each of the samples, initial QC was performed, while the metric measurements are provided as Supplementary Material S1.
Tumour DNA and matched germline DNA samples derived from 30 patients with primary BC were sequenced using the GMCK Solid Cancer Panel. The variant caller algorithm VarDict was used for mutational pattern analysis and two de novo mutational signatures (Fig. 3). Before excluding variants below 5% variant allele frequency, VarDict detected 3,665,340 somatic SNVs in total, with very high proportion of C>T substitutions and very low proportion for all other point mutation across the samples (Fig. 3a). After excluding variants below 5% variant allele frequency, VarDict detected 153,999 somatic SNVs in total across the samples (Fig. 3b). The relative contribution of C>T substitutions was substantially lower after excluding variants below 5% variant allele frequency compared to prior filtering. The opposite effect was observed for all other point mutation types. The two de novo mutational signatures were also extracted based on non-negative matrix factorization for the VarDict caller, before (Fig. 3c) and after excluding variants below 5% variant allele frequency (Fig. 3d). Based on the mutation pattern analysis, before filtering samples had the highest proportion of C>T substitutions compared to post-filtering.
Substitution frequency barplots across samples show the relative contribution of the indicated mutation types to the point mutation spectrum for VarDict caller, (a) before and (b) after excluding variants below 5% variant allele frequency32. Before filtering, samples show high proportion of C>T substitutions. Bars depict the mean relative contribution of each mutation type over all the samples. The total number of somatic point mutations across samples, after tumour-normal paired analysis of 30 patients, is indicated for VarDict caller. Two de novo mutational signatures extracted based on non-negative matrix factorization (NMF) using VarDict caller, (c) before and (d) after excluding variants below 5% variant allele frequency. Substitution frequency barplots depict the mean relative contribution of each mutation type over all the samples, after tumour-normal paired analysis of 30 patients, is indicated for VarDict caller. Overall, the mutation pattern analysis show high proportion of C>T substitutions before filtering.
Performance of the simultaneous DNA/RNA extraction protocol and gene expression profiling in archival FFPE clinical samples
Gene expression analysis of the RNA extracted from two patients enrolled in the SBG2004-1 clinical trial was performed using nCounter® Breast Cancer 360™ gene panel. The RNA yield was adequate for both samples, while 300 ng input was required for running the assay. Standardized quality assurance metrics calculation and data pre-processing revealed no QC flags or other performance issues. Moreover, all gene transcript probes exhibit the background threshold which was determined by the negative control probes included in the BC360 panel. The gene expression distribution of the top 20 endogenous genes and negative controls is illustrated in box plots (provided as Supplementary Material S1), while all QC metrics concerning the nCounter BC360 panel are summarized in a table (provided as Supplementary Material S1). All generated raw and pre-processed gene expression data for the 776 gene targets are listed as Supplementary Material S1.
Discussion
High-throughput downstream application technologies represent powerful tools for understanding cancer biology and developing clinically valuable biomarkers and targeted therapies. Given that archival FFPE material is often the only available tissue source for translational research studies, its challenging nature can limit the performance of such genomic and transcriptomic methods. The present study focusing on archival FFPE BC tissue, represents a multi-level approach including: (i) a systematic literature search of the studies comparing FF and FFPE-matched BC tissue patient material; (ii) experimental comparison and evaluation of three commercially available DNA/RNA extraction kits and (iii) feasibility study of the simultaneous DNA/RNA extraction protocol using archival tissue from patients enrolled in a randomized trial for targeted DNA-sequencing and gene expression analysis.
A high degree of concordance between FF and FFPE-matched tissue samples derived from patients with BC was observed for both RNA and DNA analyses according to the published literature. The highest degree of concordance among different platforms for gene expression analysis was reported with nCounter, a hybridization-based platform (Nanostring), showing credibility for low yielded RNA samples45,48,54,59,61,62. For genomic analysis, although a few studies reported comparison metrics on the performance of DNA-seq between FF and FFPE-matched material, the degree of concordance as reported in individual studies remained high. Of note, this concordance is constantly improving over the years, indicating the evolution of already available and dynamic development of new powerful sequencing technologies and analysis tools.
In our study, the simultaneous DNA/RNA extraction method (AllPrep) compared to RNA-only (RNeasy) and DNA-only (QIAamp) extraction methods performed similarly in all three initial QC metrics, including purity assessment, quantification, and integrity estimation of the nucleic acids. The initial QC metrics of the nucleic acids extracted from FFPE material are pertinent. Despite the purity ratios, \(A_{260}\)/\(A_{280}\) and \(A_{260}\)/\(A_{230}\), and the relatively low integrity of the nucleic acids, a more important factor when it comes to FFPE sample selection for certain technique remains the yield assessment so as to calculate the input amount. For all three silica-based extraction methods, DNA yield based on Nanodrop quantification was up to 3-fold higher compared to Qubit, while a lower discrepancy was noticed for RNA yield. The fluorescence-based nucleic acid quantification method (i.e., Qubit fluorometer) seems to be more sensitive compared to spectrophotometer-based methods, possibly due to the selective binding mode of the fluorescent dyes on different nucleic acids. Spectrophotometer’s nucleic acid quantification algorithms are more prone to overestimate the nucleic acid yield due to their disadvantage in distinguishing ribonucleotide and deoxyribonucleotide formations. In addition to the DIN and RIN\(^{e}\) values, the DV\(_{200}\) value has been proposed as an additional quality metric for the estimation of RNA integrity76. A weak correlation between different RNA integrity metrics is noticed in low-yield FFPE samples compared to FF77, calling both RIN\(^{e}\) and DV\(_{200}\) metrics in question regarding the qualification for downstream applications78.
In order to demonstrate the feasibility of the simultaneous DNA/RNA extraction method (AllPrep) in the context of an early breast cancer clinical trial, targeted DNA sequencing was performed using a hybrid capture-based panel for genomic DNA extracted from tumour-rich surgical FFPE blocks and for matched germline DNA extracted from whole peripheral blood. Although the libraries were constructed successfully, the bioinformatic analysis of the NGS data had to be optimised in order to overcome potential artefacts due to the challenging nature of the FFPE samples. The high C:G>T:A transitions appeared to be an artefact of FFPE samples due to cytosine deamination30. To better interpret the sequencing data, mutation pattern analysis was performed using the VarDict variant caller algorithm, before and after excluding variants below 5% variant allele frequency. The latter is a previously suggested post-filtering strategy32 to remove the high proportion of C>T/G>A. We have achieved similar relative contributions of the indicated mutation types to the point mutation spectrum, as previously reported32. Several pre-sequencing method approaches have been proposed to improve the accuracy of DNA sequencing data, such as the pretreatment with several DNA glycosylases79,80,81,82,83.
Tissue morphology is better preserved in FFPE biospecimens, improving pathology evaluation compared to FF. Additionally, long-term storage in non-sterile conditions requires less maintenance and fewer resources compared to FF. On the other hand, nucleic acids extracted from FFPE are often degraded and prone to artefacts compared to FF2,3. Therefore, the collection of both FF and FFPE tissue material is preferable, in order to minimize pre-analytical variability of prospectively collected tissues84.
One of the limitations of this study is publication bias which may have led to overestimation of the correlation between FF and FFPE samples. Another limitation of the systematic review is that it only included studies that reported on specific platforms and methods. Therefore, nucleic acid analysis technique platforms like other PCR-based methods (e.g., qPCR, RT-qPCR, hydrolysis of pre-existing sequences TaqMan®), DNA methylation analysis methods (e.g., quantitative bisulfite pyrosequencing), chromatin immunoprecipitation sequencing (i.e., ChIP-Seq) were not included in our study. Moreover, the studies reported performance comparisons between FF and FFPE-matched nucleic acid material, often descriptively and at different levels, making it difficult to further analyze and draw certain conclusions. For the pre-analytical performance part, a limitation is that the compared nucleic acid extraction kits are of the same silica gel membrane technology, while no other extraction kits based on paramagnetic particles or glass fibers were tested78,85. Further, due to the low sample size, no significant differences in QC metrics were obtained when comparing the AllPrep with the other two kits. The feasibility study workflow, using the simultaneous DNA/RNA extraction method on archival surgical FFPE material was demonstrated by the performance of the extracted DNA using the targeted DNA GMCK panel for thirty patients, while its performance for the extracted RNA was demonstrated using nCounter BC360 panel only for two patients.
Conclusion
The present study highlights the importance of optimising the pre-analytical variables and the QC metrics to evaluate better the yield, purity and integrity of the nucleic acids extracted from FFPE samples. The initial QC would be of particular interest when applying costly high-throughput platforms on large-scale clinical trial material. Overall, we demonstrated the feasibility of simultaneous DNA and RNA extraction method from FFPE material using downstream applications, which are also highlighted in the systematic literature search part with high to excellent concordance. We also address the need in bioinformatics to exclude variants below 5% variant allele frequency to overcome FFPE-induced artefacts. This feasibility study-workflow, including targeted DNA NGS and gene expression analysis, serves as a pilot study for larger trials. In conclusion, our findings might provide input to translational studies where FFPE material is the only available patient tumour tissue resource.
Data availability
The raw and pre-processed gene expression data for the 776 gene targets generated in this study are included in the Supplementary Material. The raw DNA sequencing data files that support the findings of this study can be obtained from the corresponding author (D.S.) upon reasonable request, provided that the intended use aligns with the ethics approval and the informed consent signed by the trial participants.
References
Harbeck, N. et al. Breast cancer. Nat. Rev. Dis. Primers 5, 1–31. https://doi.org/10.1038/s41572-019-0111-2 (2019).
Greytak, S. R., Engel, K. B., Bass, B. P. & Moore, H. M. Accuracy of molecular data generated with ffpe biospecimens: Lessons from the literature. Can. Res. 75, 1541–1547. https://doi.org/10.1158/0008-5472.CAN-14-2378 (2015).
Bass, B. P., Engel, K. B., Greytak, S. R. & Moore, H. M. A review of preanalytical factors affecting molecular, protein, and morphological analysis of formalin-fixed, paraffin-embedded (ffpe) tissue: How well do you know your ffpe specimen?. Arch. Pathol. Lab. Med. 138, 1520–1530. https://doi.org/10.5858/ARPA.2013-0691-RA (2014).
Bonin, S. & Stanta, G. Nucleic acid extraction methods from fixed and paraffin-embedded tissues in cancer diagnostics. Expert Rev. Mol. Diagn. 13, 271–282. https://doi.org/10.1586/ERM.13.14 (2013).
Romero-Pérez, L. & Grünewald, T. G. Tissue preservation and ffpe samples: Optimized nucleic acids isolation in ewing sarcoma. Methods Mol. Biol. 2226, 27–38. https://doi.org/10.1007/978-1-0716-1020-6_3 (2021).
Fox, C. H., Johnson, F. B., Whiting, J. & Roller, P. P. Formaldehyde fixation. J. Histochem. Cytochem. 33, 845–853. https://doi.org/10.1177/33.8.3894502 (1985).
Hoffman, E. A., Frey, B. L., Smith, L. M. & Auble, D. T. Formaldehyde crosslinking: A tool for the study of chromatin complexes. J. Biol. Chem. 290, 26404–26411. https://doi.org/10.1074/JBC.R115.651679 (2015).
Hewitt, S. M. et al. Tissue handling and specimen preparation in surgical pathology: Issues concerning the recovery of nucleic acids from formalin-fixed, paraffin-embedded tissue. Arch. Pathol. Lab. Med. 132, 1929–1935. https://doi.org/10.5858/132.12.1929 (2008).
Thavarajah, R., Mudimbaimannar, V. K., Elizabeth, J., Rao, U. K. & Ranganathan, K. Chemical and physical basics of routine formaldehyde fixation. J. Oral Maxillofac. Pathol. 16, 400–405. https://doi.org/10.4103/0973-029X.102496 (2012).
Carithers, L. J. et al. The biospecimen preanalytical variables program: A multiassay comparison of effects of delay to fixation and fixation duration on nucleic acid quality. Arch. Pathol. Lab. Med. 143, 1106–1118. https://doi.org/10.5858/ARPA.2018-0172-OA (2019).
Greytak, S. R., Engel, K. B. & Moore, H. M. Maximizing the utility of archival formalin-fixed paraffin-embedded blocks for nucleic acid analysis. https://home.liebertpub.com/bio16, 245–246. https://doi.org/10.1089/BIO.2018.29042.SJG (2018).
Page, M. J. et al. The prisma 2020 statement: an updated guideline for reporting systematic reviews. BMJ 372. https://doi.org/10.1136/BMJ.N71 (2021).
Bramer, W. & Bain, P. Updating search strategies for systematic reviews using endnote. J. Med. Library Assoc. 105, 285–289. https://doi.org/10.5195/JMLA.2017.183 (2017).
Bramer, W. M., Giustini, D., de Jonge, G. B., Holland, L. & Bekhuis, T. De-duplication of database search results for systematic reviews in endnote. J. Med. Library Assoc. 104, 240–243. https://doi.org/10.5195/JMLA.2016.24 (2016).
Margolin, S. et al. A randomised feasibility/phase ii study (sbg 2004–1) with dose-dense/tailored epirubicin, cyclophoshamide (ec) followed by docetaxel (t) or fixed dosed dose-dense ec/t versus t, doxorubicin and c (tac) in node-positive breast cancer. Acta Oncol. 50, 35–41. https://doi.org/10.3109/0284186X.2010.535847 (2011).
Matikas, A. et al. Long-term safety and survival outcomes from the scandinavian breast group 2004–1 randomized phase ii trial of tailored dose-dense adjuvant chemotherapy for early breast cancer. Breast Cancer Res. Treat. 168, 349–355. https://doi.org/10.1007/S10549-017-4599-4 (2018).
Zerdes, I. et al. Interplay between copy number alterations and immune profiles in the early breast cancer scandinavian breast group 2004-1 randomized phase ii trial: results from a feasibility study. NPJ Breast Cancer 7, 1–11. https://doi.org/10.1038/s41523-021-00352-3 (2021).
Foukakis, T. et al. Effect of tailored dose-dense chemotherapy vs standard 3-weekly adjuvant chemotherapy on recurrence-free survival among women with high-risk early breast cancer: a randomized clinical trial. JAMA 316, 1888–1896 (2016).
Wilfinger, W. W., Mackey, K. & Chomczynski, P. Effect of ph and ionic strength on the spectrophotometric assessment of nucleic acid purity. Biotechniques 22, 474–481. https://doi.org/10.2144/97223ST01 (1997).
Asl, H. F. Balsamic: A bioinformatic analysis pipeline for somatic mutations in cancer [online] (2019). Available online at: https://github.com/Clinical-Genomics/BALSAMIC.
Andrews, S. Fastqc: A quality control tool for high throughput sequence data [online] (2010). Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: An ultra-fast all-in-one fastq preprocessor. Bioinformatics 34, i884–i890. https://doi.org/10.1093/BIOINFORMATICS/BTY560 (2018).
Li, H. & Durbin, R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25, 1754–1760. https://doi.org/10.1093/BIOINFORMATICS/BTP324 (2009).
Li, H. et al. The sequence alignment/map format and samtools. Bioinformatics 25, 2078–2079. https://doi.org/10.1093/BIOINFORMATICS/BTP352 (2009).
Li, H. & Barrett, J. A statistical framework for snp calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993. https://doi.org/10.1093/BIOINFORMATICS/BTR509 (2011).
Picard toolkit.” 2018. broad institute, github repository. available online at: http://broadinstitute.github.io/picard/ (2018).
Ewels, P., Magnusson, M., Lundin, S. & Käller, M. Multiqc: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048. https://doi.org/10.1093/BIOINFORMATICS/BTW354 (2016).
Lai, Z. et al. Vardict: A novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res. 44, e108–e108. https://doi.org/10.1093/NAR/GKW227 (2016).
McLaren, W. et al. The ensembl variant effect predictor. Genome Biol. 17, 1–14. https://doi.org/10.1186/S13059-016-0974-4 (2016).
Do, H. & Dobrovic, A. Sequence artifacts in dna from formalin-fixed tissues: Causes and strategies for minimization. Clin. Chem. 61, 64–71. https://doi.org/10.1373/CLINCHEM.2014.223040 (2015).
Spencer, D. H. et al. Comparison of clinical targeted next-generation sequence data from formalin-fixed and fresh-frozen tissue specimens. J. Mol. Diagn. 15, 623–633. https://doi.org/10.1016/j.jmoldx.2013.05.004 (2013).
Bhagwate, A. V. et al. Bioinformatics and dna-extraction strategies to reliably detect genetic variants from ffpe breast tissue samples. BMC Genomics 20, 1–10. https://doi.org/10.1186/S12864-019-6056-8 (2019).
Chen, S. et al. A genome-wide mutational constraint map quantified from variation in 76,156 human genomes. bioRxiv 2022.03.20.485034. https://doi.org/10.1101/2022.03.20.485034 (2022).
Tate, J. G. et al. Cosmic: The catalogue of somatic mutations in cancer. Nucleic Acids Res. 47, D941–D947. https://doi.org/10.1093/NAR/GKY1015 (2019).
Manders, F. et al. Mutationalpatterns: The one stop shop for the analysis of mutational processes. BMC Genomics 23, 1–18. https://doi.org/10.1186/S12864-022-08357-3 (2022).
Bibikova, M. et al. Quantitative gene expression profiling in formalin-fixed, paraffin-embedded tissues using universal bead arrays. Am. J. Pathol. 165, 1799–1807. https://doi.org/10.1016/S0002-9440(10)63435-9 (2004).
Loudig, O. et al. Molecular restoration of archived transcriptional profiles by complementary-template reverse-transcription (ct-rt). Nucleic Acids Res. 35, e94. https://doi.org/10.1093/NAR/GKM510 (2007).
Duenwald, S. et al. Development of a microarray platform for ffpet profiling: application to the classification of human tumors. J. Transl. Med. 7. https://doi.org/10.1186/1479-5876-7-65 (2009).
Waddell, N. et al. Gene expression profiling of formalin-fixed, paraffin-embedded familial breast tumours using the whole genome-dasl assay. J. Pathol. 221, 452–461. https://doi.org/10.1002/PATH.2728 (2010).
Kibriya, M. G. et al. Analyses and interpretation of whole-genome gene expression from formalin-fixed paraffin-embedded tissue: an illustration with breast cancer tissues. BMC Genomics11. https://doi.org/10.1186/1471-2164-11-622 (2010).
Mittempergher, L. et al. Gene expression profiles from formalin fixed paraffin embedded breast cancer tissue are largely comparable to fresh frozen matched tissue. PloS One 6. https://doi.org/10.1371/JOURNAL.PONE.0017163 (2011).
Morrogh, M. et al. Differentially expressed genes in window trials are influenced by the wound-healing process: Lessons learned from a pilot study with anastrozole. J. Surg. Res. 176, 121–132. https://doi.org/10.1016/J.JSS.2011.05.058 (2012).
Meng, W. et al. Comparison of microrna deep sequencing of matched formalin-fixed paraffin-embedded and fresh frozen cancer tissues. PLoS ONE 8, e64393. https://doi.org/10.1371/JOURNAL.PONE.0064393 (2013).
Li, S. et al. Deep sequencing reveals small rna characterization of invasive micropapillary carcinomas of the breast. Breast Cancer Res. Treat. 136, 77–87. https://doi.org/10.1007/S10549-012-2166-6 (2012).
Norton, N. et al. Gene expression, single nucleotide variant and fusion transcript discovery in archival material from breast tumors. PLoS ONE 8, e81925. https://doi.org/10.1371/JOURNAL.PONE.0081925 (2013).
Sapino, A. et al. Mammaprint molecular diagnostics on formalin-fixed, paraffin-embedded tissue. J. Mol. Diagn. JMD 16, 190–197. https://doi.org/10.1016/J.JMOLDX.2013.10.008 (2014).
Nishio, M. et al. 72-gene classifier for predicting prognosis of estrogen receptor-positive and node-negative breast cancer patients using formalin-fixed, paraffin-embedded tumor tissues. Clin. Breast Cancer 14. https://doi.org/10.1016/J.CLBC.2013.11.006 (2014).
Andrade, V. P. et al. Gene expression profiling of lobular carcinoma in situ reveals candidate precursor genes for invasion. Mol. Oncol. 9, 772–782. https://doi.org/10.1016/J.MOLONC.2014.12.005 (2015).
Zhao, W. et al. Comparison of rna-seq by poly (a) capture, ribosomal rna depletion, and dna microarray for expression profiling. BMC Genom. textbf15. https://doi.org/10.1186/1471-2164-15-419 (2014).
Musella, V. et al. Use of formalin-fixed paraffin-embedded samples for gene expression studies in breast cancer patients. PloS One 10. https://doi.org/10.1371/JOURNAL.PONE.0123194 (2015).
Beumer, I. et al. Equivalence of mammaprint array types in clinical trials and diagnostics. Breast Cancer Res. Treat. 156, 279. https://doi.org/10.1007/S10549-016-3764-5 (2016).
Jovanović, B. et al. Comparison of triple-negative breast cancer molecular subtyping using rna from matched fresh-frozen versus formalin-fixed paraffin-embedded tissue. BMC Cancer17. https://doi.org/10.1186/S12885-017-3237-1 (2017).
Loudig, O. et al. Evaluation and adaptation of a laboratory-based cdna library preparation protocol for retrospective sequencing of archived micrornas from up to 35-year-old clinical ffpe specimens. Int. J. Mol. Sci. 18, 627. https://doi.org/10.3390/IJMS18030627 (2017).
Yamaguchi, S. et al. Molecular and clinical features of the tp53 signature gene expression profile in early-stage breast cancer. Oncotarget 9, 14193–14206. https://doi.org/10.18632/ONCOTARGET.24447 (2018).
Jose, V. et al. Feasibility of developing reliable gene expression modules from ffpe derived rna profiled on affymetrix arrays. PLoS ONE 13, e0203346. https://doi.org/10.1371/JOURNAL.PONE.0203346 (2018).
Loudig, O., Liu, C., Rohan, T. & Ben-Dov, I. Z. Retrospective microrna sequencing: Complementary dna library preparation protocol using formalin-fixed paraffin-embedded rna specimens. J. Vis. Exp. JoVE 2018, 57471. https://doi.org/10.3791/57471 (2018).
Wrzeszczynski, K. O. et al. Analytical validation of clinical whole-genome and transcriptome sequencing of patient-derived tumors for reporting targetable variants in cancer. J. Mol. Diagn. JMD 20, 822–835. https://doi.org/10.1016/J.JMOLDX.2018.06.007 (2018).
Li, J., Fu, C., Speed, T. P., Wang, W. & Symmans, W. F. Accurate rna sequencing from formalin-fixed cancer tissue to represent high-quality transcriptome from frozen tissue. JCO Precis. Oncol. 1–9, 2018. https://doi.org/10.1200/PO.17.00091 (2018).
Stewart, R. L. et al. A multigene assay determines risk of recurrence in patients with triple-negative breast cancer. Can. Res. 79, 3466–3478. https://doi.org/10.1158/0008-5472.CAN-18-3014 (2019).
Marczyk, M. et al. The impact of rna extraction method on accurate rna sequencing from formalin-fixed paraffin-embedded tissues. BMC Cancer 19, 1–12. https://doi.org/10.1186/S12885-019-6363-0 (2019).
Turnbull, A. K. et al. Unlocking the transcriptomic potential of formalin-fixed paraffin embedded clinical tissues: Comparison of gene expression profiling approaches. BMC Bioinformatics 21, 1–10. https://doi.org/10.1186/S12859-020-3365-5 (2020).
Sun, J. et al. Development of malignancy-risk gene signature assay for predicting breast cancer risk. J. Surg. Res. 245, 153–162. https://doi.org/10.1016/J.JSS.2019.07.021 (2020).
Lau, R. et al. Technical validity of a customized assay of sensitivity to endocrine therapy using sections from fixed breast cancer tissue. Clin. Chem. 66, 934–945. https://doi.org/10.1093/CLINCHEM/HVAA105 (2020).
Bergeron, D. et al. Rna-seq for the detection of gene fusions in solid tumors: development and validation of the jax fusionseq\(^{{\rm TM}}\) 2.0 assay. J. Mol. Med. (Berlin, Germany)100, 323–335. https://doi.org/10.1007/S00109-021-02149-0 (2022).
Liu, Y. et al. Quality control recommendations for rnaseq using ffpe samples based on pre-sequencing lab metrics and post-sequencing bioinformatics metrics. BMC Med. Genom. 15. https://doi.org/10.1186/S12920-022-01355-0 (2022).
Hilmi, M., Armenoult, L., Ayadi, M. & Nicolle, R. Whole-transcriptome profiling on small ffpe samples: Which sequencing kit should be used?. Curr. Issues Mol. Biol. 44, 2186–2193. https://doi.org/10.3390/CIMB44050148 (2022).
Marczyk, M. et al. Assessment of stained direct cytology smears of breast cancer for whole transcriptome and targeted messenger rna sequencing. Cancer Cytopathol. 131, 289–299. https://doi.org/10.1002/CNCY.22679 (2023).
MacConaill, L. E. et al. Profiling critical cancer gene mutations in clinical tumor samples. PloS One 4. https://doi.org/10.1371/JOURNAL.PONE.0007887 (2009).
Schweiger, M. R. et al. Genome-wide massively parallel sequencing of formaldehyde fixed-paraffin embedded (ffpe) tumor tissues for copy-number- and mutation-analysis. PloS One 4. https://doi.org/10.1371/JOURNAL.PONE.0005548 (2009).
Bourgon, R. et al. High-throughput detection of clinically relevant mutations in archived tumor samples by multiplexed pcr and next-generation sequencing. Clin. Cancer Res. 20, 2080–2091. https://doi.org/10.1158/1078-0432.CCR-13-3114 (2014).
Munchel, S. et al. Targeted or whole genome sequencing of formalin fixed tissue samples: potential applications in cancer genomics. Oncotarget 6, 25943–25961. https://doi.org/10.18632/ONCOTARGET.4671 (2015).
Martelotto, L. G. et al. Whole-genome single-cell copy number profiling from formalin-fixed paraffin-embedded samples. Nat. Med. 23, 376. https://doi.org/10.1038/NM.4279 (2017).
Robbe, P. et al. Clinical whole-genome sequencing from routine formalin-fixed, paraffin-embedded specimens: pilot study for the 100,000 genomes project. Genet. Med. 20, 1196–1205. https://doi.org/10.1038/GIM.2017.241 (2018).
Nachmanson, D. et al. Mutational profiling of micro-dissected pre-malignant lesions from archived specimens. BMC Med. Genom. 13. https://doi.org/10.1186/S12920-020-00820-Y (2020).
Wei, L., Dugas, M. & Sandmann, S. Simffpe and filterffpe: improving structural variant calling in ffpe samples. GigaScience 10. https://doi.org/10.1093/GIGASCIENCE/GIAB065 (2021).
Wimmer, I. et al. Systematic evaluation of rna quality, microarray data reliability and pathway analysis in fresh, fresh frozen and formalin-fixed paraffin-embedded tissue samples. Sci. Rep. 8, 1–17. https://doi.org/10.1038/s41598-018-24781-6 (2018).
Walker, J. E. et al. Measuring up: A comparison of tapestation 4200 and bioanalyzer 2100 as measurement tools for rna quality in postmortem human brain samples. Int. J. Mol. Sci. 24, 13795 (2023).
Landolt, L., Marti, H.-P., Beisland, C., Flatberg, A. & Eikrem, O. S. Rna extraction for rna sequencing of archival renal tissues. Scand. J. Clin. Lab. Invest. 76, 426–434 (2016).
Steiert, T. A. et al. A critical spotlight on the paradigms of ffpe-dna sequencing. Nucleic Acids Res. 51, 7143. https://doi.org/10.1093/NAR/GKAD519 (2023).
Berra, C. M. et al. Use of uracil-dna glycosylase enzyme to reduce dna-related artifacts from formalin-fixed and paraffin-embedded tissues in diagnostic routine. Appl. Cancer Res. 39, 1–6. https://doi.org/10.1186/S41241-019-0075-2 (2019).
Do, H. et al. Reducing artifactual egfr t790m mutations in dna from formalin-fixed paraffin-embedded tissue by use of thymine-dna glycosylase. Clin. Chem. 63, 1506–1514. https://doi.org/10.1373/CLINCHEM.2017.271932 (2017).
Bessho, T. et al. Repair of 8-hydroxyguanine in dna by mammalian n-methylpurine-dna glycosylase. Proc. Natl. Acad. Sci. U.S.A. 90, 8901–8904. https://doi.org/10.1073/PNAS.90.19.8901 (1993).
Xiong, K. et al. Duplex-repair enables highly accurate sequencing, despite dna damage. Nucleic Acids Res. 50. https://doi.org/10.1093/NAR/GKAB855 (2022).
Engel, K. B., Vaught, J. & Moore, H. M. National cancer institute biospecimen evidence-based practices: a novel approach to pre-analytical standardization. Biopreserv. Biobank. 12, 148–150 (2014).
McDonough, S. J. et al. Use of ffpe-derived dna in next generation sequencing: Dna extraction methods. PLoS ONE 14, e0211400 (2019).
Acknowledgements
Alexios Matikas is supported by the Swedish Cancer Society (Cancerfonden) Junior Clinical Investigator award. Ioannis Zerdes is supported by the Region Stockholm (clinical postdoctorial appointment, FoUI 977295), the Swedish Society of Oncology postdoctoral grant and the Iris, Stig och Gerry Castenbäcks foundation We would like to thank Anna Gellerbring and Keyvan Elhami from the Facility of Clinical Genomics at Science for Life Laboratory, Stockholm, Sweden for their contribution to this study. We would also like to thank Gun Brit Knutssoen and Narcisa Hannerz, librarians at Karolinska Institutet University Library (KIB) for their contribution to the literature search. We would like to acknowledge the contribution of Annika Larsson from the KIGene core facility at Karolinska Institutet during the experimental part of the Nanosting BC360 platform and Nikos Tsiknakis for preparing the manuscript in LaTeX format.
Funding
Open access funding provided by Karolinska Institute. This study was supported by grants from Region Stockholm, Karolinska Institutet including Cancer Research KI, the Swedish Cancer Society, the Research Funds at Radiumhemmet.
Author information
Authors and Affiliations
Contributions
Conceptualization: D.S., T.F., A.M., I.Z.; Clinical samples: D.S., S.A., J.B., I.Z.; Data curation: D.S., E.S., A.M., I.Z.; Formal analysis: D.S., E.S., A.M., I.Z.; Data analysis: D.S., E.S., A.M., I.Z.; Interpretation of data: D.S., E.S., A.M., I.Z.; Funding acquisition: T.F., A.M., I.Z.; Investigation: D.S., E.S., A.M., I.Z.; Methodology: D.S., E.S., S.A., V.W., J.H., A.M., I.Z.; Project administration: D.S., A.M., I.Z.; Software: D.S., E.S.; Supervision: A.M., I.Z.; Visualization: D.S., E.S., V.W.; Writing: D.S., E.S., V.W., T.F., A.M., I.Z.; manuscript review and approval: All authors
Corresponding author
Ethics declarations
Competing interests
Alexios Matikas: consultancy or speaker (no personal fees): Roche, Veracyte, Seagen. Research funding paid to institution: AstraZeneca, Merck, Novartis; Valtteri Wirta: speaker’s honoraria and reimbursement of travel expenses from Illumina; Johan Hartman: speaker’s honoraria or advisory board remunerations from Roche, Novartis, Pfizer, EliLilly, MSD and institutional research support from Roche, AstraZeneca and Novartis. Co-founder and shareholder of Stratipath AB; Jonas Bergh: Research grants from Amgen, AstraZeneca, Bayer, Merck, Pfizer, Roche and Sanofi-Aventis to Karolinska Institutet and/or University Hospital. No personal payments. Co-author on a chapter on “Prognostic and Predictive factors in early, non-metastatic breast cancer” in UpToDate. Honoraria to Asklepios Medicin HB. Stocks in Stratipath AB, a company involved in Al-based diagnostics for breast cancer. Chairperson for Coronis and Asklepios Cancer Research HB. Honoraria from Roche and AstraZeneca for chairmanship and lectures at scientific meetings and consultations for Stratipath AB; Theodoros Foukakis: Financial Interests, Institutional, Invited Speaker: Roche, AstraZeneca, Gilead Sciences. Financial Interests, Personal, Advisory Board: Novartis, Veracyte, Exact Sciences, Affibody. Financial Interests, Personal, Invited Speaker: Pfizer. Financial Interests, Personal, Royalties, Authorship of two chapters in UpToDate: Wolters Kluwer. Financial Interests, Institutional, Coordinating PI, Clinical trial support (research grant and study drug): Pfizer. Financial Interests, Institutional, Sponsor and Coordinating PI, International co-PI of academic trial ARIADNE (EU CT: 2022-501504-95-00): AstraZeneca, Novartis, Veracyte; All other authors have no conflicts of interest to disclose.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Salgkamis, D., Sifakis, E.G., Agartz, S. et al. Systematic review and feasibility study on pre-analytical factors and genomic analyses on archival formalin-fixed paraffin-embedded breast cancer tissue. Sci Rep 14, 18275 (2024). https://doi.org/10.1038/s41598-024-69285-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-69285-8
- Springer Nature Limited