Abstract
The genome is constantly exposed to DNA damage from endogenous and exogenous sources. Fine modulation of DNA repair, chromatin remodeling, and transcription factors is necessary for protecting genome integrity, but the precise mechanisms are still largely unclear. We found that after ionizing radiation (IR), global trimethylation of histone H3 at lysine 4 (H3K4me3) was decreased at an early (5 min) post-IR phase but increased at an intermediate (180 min) post-IR phase in both human and mouse hematopoietic cells. We demonstrated that PTIP, a component of the MLL histone methyltransferase complex, is required for H3K4me3 upregulation in the intermediate post-IR phase and promotes cell cycle arrest by epigenetically inducing a cell cycle inhibitor, PRDM1. In addition, we found that PTIP expression is specifically downregulated in acute myeloid leukemia patients. These findings collectively suggest that the PTIP-PRDM1 axis plays an essential role in proper DNA damage response and its deregulation contributes to leukemogenesis.
Similar content being viewed by others
Introduction
Cells have evolved highly orchestrated signaling pathways to repair DNA damage, since accumulation of DNA damage increases the risk of cancer1. In the DNA damage response (DDR), cells recognize lesions and transiently arrest cell cycle progression to allow time for repair2. DDR leads to cell cycle arrest, DNA repair, as well as apoptosis3 and prolonged cell cycle arrest following DNA damage is essential for effective DNA repair4. DNA double-strand breaks (DSBs) are the most cytotoxic type of DNA damage that occurs following DNA replication, exposure to ionizing radiation (IR), and treatment with chemotherapeutic drugs5. DSBs are repaired by either non-homologous end joining (NHEJ) or homologous recombination (HR)6. 53BP1 plays central roles in the NHEJ pathway—it is recruited to DSBs, protects DNA ends from excessive resection, and promotes end-joining of DNA ends7.
PTIP (PAX transcription activation domain interacting protein, also known as PAXIP1) contains multiple BRCA1 C-terminus (BRCT) domains that can interact with transcription factors and it functions as an adaptor protein downstream of p53-binding protein 1 (53BP1)8,9. PTIP is recruited by phosphorylated 53BP1 to promote the NHEJ pathway and PTIP-deficient cells are sensitive to DNA damage, indicating that PTIP is directly involved in DNA repair via formation of the 53BP1 complex8,10,11,12. On the other hand, PTIP functions as a component of the mixed-lineage leukemia (MLL) histone methyltransferase complexes that catalyze trimethylated histone H3 at lysine 4 (H3K4me3), a mark associated with gene activation13,14. Indeed, PTIP is required for transcriptional activation via epigenetic regulation, such as peroxisome proliferator-activated receptor gamma (PPARγ) activation in preadipocytes and osteoclasts15 and sphingosine-1-phosphate receptor 1 (S1PR1) in T lymphocyte egress16. Moreover, PTIP is associated with Igh germline transcription in B cell class switch recombination (CSR) by regulating H3K4me3 and promotes CSR17,18,19. Therefore, it is strongly suggested that PTIP contributes to cellular homeostasis through different functional properties.
Studies reported that methylation of histones at certain sites is associated with accessibility and recruitment of repair factors. Indeed, H3K4me3 and methylated H3K36 were found to promote the DDR pathway by recruiting repair factors20,21,22. In addition, H3K27me3, a gene repression mark, also promotes the NHEJ pathway through interaction with FANCD2, a key player in the DDR pathway23. While methylated histone marks induce alteration of chromatin structure and recruit repair factors to the lesion, transcriptional regulation by histone modifiers is also essential for DNA repair. For instance, EZH2, the histone methyltransferase catalyzing H3K27me3, promotes DNA repair by DNA damage-induced transcriptional silencing during the early phase following DNA damage24. In addition, H3K4me3 levels are upregulated to activate DDR genes at the intermediate phase of DNA damage25.
Whether PTIP contributes to DDR not only as a component of the 53BP1 complex but also as an epigenetic factor regulating H3K4me3 levels is not known. Since a variety of mutations in DDR genes are associated with increased risk of developing leukemia26, we explored whether PTIP-mediated H3K4me3 regulation has essential roles in DDR using human leukemia cell lines and mouse hematopoietic stem-progenitor cells (HSPCs). In this study, we provide evidence that at the intermediate post-IR phase, PTIP-mediated upregulation of H3K4me3 and subsequent induction of PRDM1 (also known as BLIMP1) are necessary for DDR mediated cell cycle arrest.
Results
DNA damage by IR induced alteration of global H3K4me3 levels
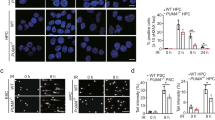
To examine whether H3K4me3 levels are altered following IR, we first performed fluorescent immunostaining and western blotting of phosphorylated H2AX (γH2AX), a marker of DNA damage, in human myeloid leukemia cell lines. In all three cell lines (HL-60, MOLM13, and K562), global H3K4me3 levels were significantly downregulated 5 min after IR but were significantly upregulated 180 min after IR in line with γH2AX accumulation, compared with 0 min (Fig. 1A–C and Supplementary Fig. 1A). Upregulated H3K4me3 levels gradually returned to baseline in a time-dependent manner (Supplementary Fig. 1B). To determine whether these observations were physiologically relevant, we performed the same experiment using primary mouse HSPCs (LSK cells; lineage [Mac-1, Gr-1, B220, Ter119, CD4 and CD8]−, c-kit+ and Sca-1+). Similar results were obtained at both time points (Fig. 1D), indicating that global H3K4me3 levels were downregulated in the early phase (5 min) following IR, likely due to DNA damage induced transcriptional silencing27, but upregulated in the intermediate phase (180 min) following IR (hereafter, referred to as early post-IR and intermediate post-IR, respectively, two timepoints in DNA repair pathway activation) in normal and leukemic hematopoietic cells.
Alteration of H3K4me3 levels after IR in human leukemia cell lines and mouse hematopoietic stem/precursors cells (HSPCs). (A-C) Immunofluorescence staining (H3K4me3, γH2AX, and Hoechst) (left panels) and relative fluorescence intensity (H3K4me3) (right panels) in irradiated human leukemia cell lines: HL-60 (A), MOLM13 (B), and K562 (C). (D) Immunofluorescence staining (H3K4me3 and Hoechst) (left panels) and relative fluorescence intensity (H3K4me3) (right panels) in mouse LSK (lineage [Mac-1, Gr-1, B220, Ter119, CD4 and CD8]−, c-kit+ and Sca-1+) cells. The relative intensities at 5 min after ionizing radiation (IR) and 180 min after IR are compared with 0 min after IR. The mean values are indicated as bars (n = 80). ***p < 0.001.
PTIP induces upregulation of global H3K4me3 levels at the intermediate phase after IR
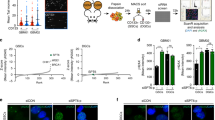
Since PTIP mediates H3K4 methylation as part of a histone methyltransferase complex and the cell lines we tested do not possess mutations or deletions at the PTIP gene (COSMIC, https://cancer.sanger.ac.uk/cosmic), we examined whether PTIP contributes to the upregulation of H3K4me3 at intermediate post-IR. To this end, we produced PTIP-knockdown human leukemic cells and Ptip deficient mouse hematopoietic cells. For human cells, lentiviral-based control shRNA vector (shCtrl) and two independent shRNA vectors targeting PTIP (shPTIP #1 and #2) were transduced into HL-60 cells (Fig. 2A). In contrast to shCtrl-transduced HL-60 cells that exhibited significant upregulation of global H3K4me3 levels at 180 min after IR, no apparent changes were detected in shPTIP-transduced cells (Fig. 2B). The global H3K4me3 levels at 0 min after IR in shCtrl, PTIP (shPTIP #1 and #2)-transduced HL-60 cells were comparable, suggesting that PTIP deficiency may not affect H3K4me3 regulation at steady state (Fig. 2C). To obtain Ptip-deficient mouse hematopoietic cells, mice in which exon 7 was floxed (Ptipflox/flox) were crossed with ERT2Cre+ mice in which Cre was activated by tamoxifen, and Ptipflox/flox, ERT2Cre+ mice were treated with tamoxifen to generate Ptip-deficient (PtipΔ/Δ) mice (Fig. 2D). Tamoxifen-treated Ptipflox/flox, ERT2Cre− mice were used as a control (Ptip+/+). As observed in human leukemic cells, Ptip+/+ LSK cells exhibited marked upregulation of global H3K4me3 levels at 180 min after IR, which was observed to a lesser extent in PtipΔ/Δ cells (Fig. 2E). H3K4me3 upregulation in PtipΔ/Δ LSK cells was not completely diminished, potentially due to regulation of H3K4me3 by the COMPASS complex including SETD1A/B or by the COMPASS-like complex including MLL1/2. These results indicated that PTIP is required for upregulation of global H3K4me3 levels in both human and mouse hematopoietic cells at intermediate post-IR.
Alteration of H3K4me3 levels after IR in PTIP-deficient human leukemia cell lines and mouse HSPCs. (A) qPCR analysis of PTIP mRNA expression levels following introduction of shCtrl and shPTIP (#1 and #2) lentiviral vectors into HL-60 cells (mean + SD, n = 3). (B) Immunofluorescence staining (H3K4me3 and Hoechst) (left panels) and relative fluorescence intensity (H3K4me3) (right panels) in shCtrl (left), shPTIP #1 (middle), and shPTIP #2 (right) transduced HL-60 cells after IR. The relative intensity at 180 min after IR is compared with 0 min after IR for each cell line. The mean values are indicated as bars (n = 35). (C) H3K4me3 relative fluorescence intensity in shCtrl , shPTIP #1, and shPTIP #2 transduced HL-60 cells at 0 min after IR. The relative intensity at 0 min after IR is compared with shCtrl. The mean values are indicated as bars (n = 35). (D) Generation of Ptip-deficient (PtipΔ/Δ) mice. Upper panel: mice in which exon 7 was floxed (floxed allele) were crossed with ERT2Cre+ mice in which Cre was activated by tamoxifen to generate Ptipflox/flox, ERT2Cre+ mice, and exon 7 was deleted by tamoxifen administration (disrupted (Δ) allele). Lower panel: genomic PCR to detect flox allele-derived and Δ allele-derived products using bone marrow (BM) cells of tamoxifen-untreated ( −) and -treated ( +) Ptipflox/flox, ERT2Cre+ mice. (E) Immunofluorescence staining (H3K4me3 and Hoechst) (left panels) and relative fluorescence intensity (H3K4me3) (right panels) of H3K4me3 in Ptip+/+ (upper) and PtipΔ/Δ (bottom) mouse LSK cells after IR. The relative intensity at 180 min after IR is compared with 0 min after IR in each group. The mean values are indicated as bars (n = 90). *p < 0.05, ***p < 0.001.
PTIP suppresses cell cycle progression after IR via upregulation of PRDM1
A previous report identified IR-responsive genes that are associated with DDR and upregulated after IR25. We thus hypothesized that PTIP contributes to DDR through a H3K4me3-mediated transcriptional program at intermediate post-IR. To investigate the expression changes of IR-responsive genes in leukemic cells, we first performed RT-qPCR using RNA extracted from HL-60 and MOLM13 cells before and after IR. We found that the expression of PRDM1, encoding a cell cycle inhibitor28,29, was upregulated in both HL-60 and MOLM13 cells at 180 min after IR (Fig. 3A). Upregulation of PRDM1 at 180 min after IR was markedly attenuated in shPTIP-transduced HL-60 cells compared with shCtrl-transduced cells (Fig. 3B). These findings strongly suggest that PTIP contributes to upregulation of PRDM1 at intermediate post-IR. In addition, to explore the underlying transcriptional mechanisms, gene expression profiles of shCtrl- and shPTIP (#1 and #2)-transduced HL-60 cells at 0 min and 180 min after IR were examined by RNA-seq and the results were subjected to gene set enrichment analysis (GSEA) for pathway assessment (Fig. 3C and Supplementary Table. 1). In shCtrl-transduced HL-60 cells, gene sets associated with cell cycle proliferation (POSITIVE_REGULATION_OF_CELL_PROLIFERATION) were negatively enriched at 180 min after IR compared with 0 min, but no such effect was observed in shPTIP-transduced HL-60 cells (Fig. 3D, upper panels). Although no obvious changes were observed between shCtrl- and shPTIP-transduced HL-60 cells at 0 min after IR, these pathways were significantly positively enriched in shPTIP-transduced cells compared with shCtrl-transduced cells at 180 min after IR (Fig. 3D, lower panels). Following DNA damage, cell cycle checkpoints are activated and cell cycle arrest is transiently induced2,7, suggesting that PTIP contributes to IR-induced cell cycle arrest.
PTIP positively regulates PRDM1 expression after IR to arrest the cell cycle. (A) qPCR analysis of mRNA expression levels of IR-induced genes25 in HL-60 and MOLM13 cells. The expression each gene at 180 min after IR is compared with 0 min (mean + SD, n = 3). (B) qPCR analysis of PRDM1 mRNA expression in HL-60 transfected with shCtrl and shPTIP (#1 and #2) at 0 min and at 180 min after IR. The relative expression levels are compared with shCtrl at 0 min after IR (mean + SD, n = 3). (C) Schematic illustration of RNA-seq in HL-60 transfected with shCtrl and shPTIP (#1 and #2) at 0 min and 180 min after IR. (D) GSEA plots associated with cell cycle progression (POSITIVE_REGULATION_OF_CELL_PROLIFERATION, from Human MSigDB Collections, C5: ontology gene sets) in HL-60 cells transduced with shCtrl and shPTIP (#1 and #2). HL-60 cells transfected with shCtrl at 180 min after IR relative to 0 min after IR and shPTIP at 180 min after IR relative to 0 min after IR are shown (upper panels). HL-60 cells transfected with shPTIP at 0 min after IR relative to shCtrl at 0 min and shPTIP (#1 and #2) at 180 min after IR relative to shCtrl at 180 min are shown (lower panels). (E) Schematic illustration of BrdU incorporation assays in HL-60 cells after IR. (F) Flow cytometric analysis of BrdU incorporation in untreated and IR-treated HL-60 cells (mean + SD, n = 3). (G) Flow cytometric analysis of BrdU incorporation in untreated and IR treated HL-60 cells transfected with shCtrl and shPTIP (#1 and #2) (mean + SD, n = 3). (H) qPCR analysis of PRDM1 mRNA expression in HL-60 transfected with shCtrl and shPRDM1 (#1 and #2) compared with shCtrl (mean + SD, n = 3). (I) Flow cytometric analysis of BrdU incorporation in untreated or irradiated HL-60 cells transfected with shCtrl and shPRDM1 (#1 and #2) (mean + SD, n = 3). NES normalized enrichment score, FDR false discovery rate, *p < 0.05, ***p < 0.001.
To further investigate whether PTIP affects cell cycle activity via expression of PRDM1, we performed BrdU incorporation assays using IR-treated cells (Fig. 3E). We first examined the effect of IR on cell proliferation in HL-60 cells. As expected, IR-treated HL-60 cells exhibited markedly suppressed BrdU incorporation compared with untreated cells (Fig. 3F). We then performed the same assay using shCtrl- and shPTIP-transduced HL-60 cells. In contrast to shCtrl-transduced cells which exhibited significantly reduced BrdU incorporation, no such effects were detected in shPTIP-transduced cells (Fig. 3G). We further examined the effect of PRDM1 knockdown on cell proliferation following IR. We confirmed downregulation of PRDM1 with two different shRNAs (shPRDM1 #1 and #2) (Fig. 3H) and found that IR-induced cell cycle inhibition was not detected in shPRDM1-transduced cells (Fig. 3I). These findings collectively indicate that the PTIP-PRDM1 axis is essential for IR-induced cell cycle arrest.
A PTIP-containing MLL4 methyltransferase complex positively regulates H3K4me3 at the PRDM1 locus after IR
To explore the genome-wide distribution of H3K4me3, we performed CUT&RUN assays using HL-60 cells at 180 min after IR. In line with Fig. 1, genome-wide upregulation of H3K4me3 was observed in HL-60 cells after IR (Fig. 4A). We observed an increased number of H3K4me3 peaks (peak score > 3000, Supplementary Table. 2) at 180 min after IR compared with 0 min (Fig. 4B). In addition, the peaks were distributed genome-wide (Supplementary Fig. 2). The expression of H3K4me3 upregulated genes at 180 min after IR compared with 0 min, identified in Fig. 4B, was attenuated following PTIP knockdown (Fig. 4C). Motif analysis on these 1410 H3K4me3 peaks demonstrated greater enrichment for the E2F transcription factor, known to be associated with both cell cycle progression and cell cycle arrest30. Indeed, the putative target genes (± 3 kb from the transcription start sites (TSSs)) of these peaks are involved in the cell cycle pathway, which are different properties compared with the H3K4me3 peaks at 0 min (Fig. 4D,E and Supplementary Table. 3).
Epigenetic regulation of PTIP at the PRDM1 locus. (A) Box plot showing global levels of H3K4me3 in HL-60 cells at 0 min and at 180 min after IR in two replicates (Rep1 and Rep2) normalized to each IgG sample. (B) Venn diagram showing H3K4me3 peaks (peak score > 3000) in HL-60 cells at 0 min and at 180 min after IR. (C) Expression of H3K4me3 upregulated genes in HL-60 cells transfected with shCtrl and shPTIP (#1 and #2) at 180 min after IR. Genes were defined using peaks located ± 1 Kb from the TSS. (D) The top enriched transcription factor motifs from H3K4me3 peaks in HL-60 cells at 0 min after IR (top) at 180 min after IR (bottom). (E) Enrichr Pathway analysis (https://maayanlab.cloud/Enrichr/) enriched among putative target genes (± 3 kb from the TSS) from H3K4me3 peaks in HL-60 cells at 0 min after IR (top) at 180 min after IR (bottom). (F) Enrichment of H3K4me3 at the PRDM1 locus in HL-60 cells at 0 min and at 180 min after IR in the UCSC genome browser. (G) ChIP-qPCR of H3K4me3 (left), PTIP (center) and MLL4 (right) at the PRDM1 region in HL-60 cells at 0 min and 180 min after IR (mean + SD, n = 3). The primer product is indicated as blue box in Fig. 4F (H) ChIP-qPCR analysis of H3K4me3 in HL-60 cells transfected with shCtrl and shPTIP (#1 and #2) at 0 min and at 180 min after IR. (I) Schematic illustration of the dCas9-VP64 system. gRNA positions are indicated as blue boxes (#1 to #4). (J) qPCR of PRDM1 mRNA in HL-60 cells expressing dCas9-VP64 transfected with gRNA (#1 to #4). The relative expressions were compared with gRNA #4 (mean + SD, n = 3). *p < 0.05, ***p < 0.001.
To explore the direct relationship between PRDM1 expression and H3K4me3 upregulation, we analyzed whether H3K4me3 peaks accumulated at the PRDM1 locus. As expected, enrichment of H3K4me3 at PRDM1 was significantly increased downstream of the TSS in HL-60 cells at 180 min after IR compared with 0 min (Fig. 4F). In accordance with upregulation of H3K4me3, PTIP and MLL4 were also significantly enriched at this region at 180 min after IR compared with 0 min (Fig. 4G). In addition, H3K4me3 upregulation at 180 min after IR was attenuated by knockdown of PTIP (Fig. 4H). To investigate whether the regions downstream of the PRDM1 TSS are essential for transcription, dCas9-VP64 with 4 types of guide RNA (gRNA): #1 upstream of TSS, #2 and #3 putative PRDM1 regulatory regions, and #4 downstream of exon 2, were recruited with lentiviral vectors (Fig. 4I). We found that #2 and #3 regions positively regulated transcription in accordance with increased H3K4me3 (Fig. 4J). Together, these findings demonstrate that an MLL4 methyltransferase complex containing PTIP directly upregulates H3K4me3 levels downstream of the PRDM1 TSS, resulting in increased PRDM1 expression and inhibited cell cycle progression in HL-60 cells at intermediate post-IR.
PTIP is downregulated in human AML
Given that PTIP is associated with DDR directly via DNA repair and with cell cycle regulation via histone modification, PTIP is assumed to function as a tumor suppressor. To assess the role of PTIP in leukemogenesis, we compared the expression of PTIP in human leukemic samples and cell lines. We found that PTIP mRNA expression was significantly downregulated in acute myeloid leukemia (AML) samples with different karyotypes and cell lines compared with normal stem/progenitor cells (Fig. 5A–C), strongly suggesting that PTIP may play a tumor suppressive role, possibly through activation of both DDR pathways.
PTIP is downregulated in human acute myeloid leukemia (AML) and model. (A) PTIP alterations in human AML samples (n = 163) from the TCGA database (https://www.cbioportal.org). (B) Analysis of PTIP mRNA in AML cells with karyotypes of t(15;17), inv(16)/t(16;16), t(8;21), and t(11q23) translocations and the AML complex. The results of AML samples were compared with those of normal human hematopoietic stem cells (HSCs) using the Bloodspot database (https://www.fobinf.com, referred from BloodPool: AML samples vs. normal). The mean values are indicated as bars (n = 6–56). (C) qPCR analysis of PRDM1 mRNA in leukemia cells (HL-60, MOLM and K562 cells). The relative expressions were compared with cord blood derived CD34+ stem/progenitor cells as the normal cell control (mean + SD, n = 3) (D) Schematic illustration of PTIP-mediated regulation. At the intermediate phase after IR, an MLL4 complex containing PTIP upregulates global H3K4me3 levels. Expression of PRDM1, a cell cycle inhibitor, is activated by the complex via epigenetic regulation which induces cell cycle arrest to promote the DNA damage response. *p < 0.05, **p < 0.01 and ***p < 0.001.
Discussion
It has been reported that trimethylation of H3K9 and H3K27, which changes the chromatin structure to a repressive state known as heterochromatin, plays essential roles for chromatin remodeling and transcriptional silencing in DDR23,27,31. On the other hand, H3K4me3 mediated by the MLL4 methyltransferase complex containing PTIP, changes chromatin structure to an active state, called euchromatin, and is also thought to be one of the critical histone modifications for DDR. However, the mechanism underlying H3K4me3 regulation associated with PTIP in DDR has remained controversial20,21,25,32,33.
While several groups suggested the importance of H3K4me3 in DDR25,33, others argued that the demethylation of H3K4me3 positively regulates DDR20,21,32. These contradictory results raise the question as to which mark (methylated or demethylated H3K4) is beneficial for DDR. In this study, we demonstrated that global H3K4me3 levels were downregulated at early (5 min) post-IR but upregulated at intermediate (180 min) post-IR in hematopoietic cells, which is in line with the results by Rashi-Elkeles, S. et al.25 (Fig. 1). Our data support the idea that the contributions of H3K4me3 to DDR are time dependent. Indeed, treatment with JIB-04, an inhibitor of KDM5B, a demethylase of H3K4, in early post-IR induced an accumulation of H3K4me3 and impaired DNA repair pathways in non-small cell lung cancer (NSCLC) cells21. On the other hand, when NSCLC cells were treated with JIB-04 at intermediate post-IR, DNA repair pathway impairment was attenuated23. Additionally, highly compacted heterochromatin protects the genome from DNA damage but also suppresses repair efficiency and increases the mutation rate by creating a barrier to repair factor recruitment33, indicating that chromatin structure should be altered at the proper phase in DDR. These results imply that accumulation of H3K4me3 is dispensable or toxic for DDR at early post-IR but crucial for DDR at intermediate post-IR.
PTIP deficiency suppressed accumulation of H3K4me3 and promoted cell proliferation pathways at intermediate post-IR (Figs. 2, 3A), suggesting that PTIP possibly fine-tunes cell cycle progression at intermediate post-IR via H3K4me3 regulation. Our findings also highlighted that PTIP mediates expression of PRDM1, which functions as both a cell cycle inhibitor8,29 and an IRinduced protein25 by upregulating H3K4me3 in cooperation with MLL4 (Figs. 3, 4). Thus, PTIP is crucial for temporal cell cycle arrest after IR and for the DNA repair process.
In this study, we demonstrated that following DNA damage, downregulation of H3K4me3 was observed as a primary event and upregulation of H3K4me3 by an MLL4 methyltransferase complex containing PTIP occurred as a secondary event to induce cell cycle arrest via PRDM1 activation (Fig. 5D). Understanding this mechanism may also shed light on the multifaceted functions of PTIP. In addition, to address the issue that PTIP functions as a tumor suppressive factor through activation of both DDR pathways in greater detail, future studies can aim to investigate whether PtipΔ/Δ mice develop leukemia spontaneously via in vivo mutagenesis using retroviral transduction assays and exposure to a chemical carcinogen.
Materials and methods
Cells and mice
Human leukemia cell lines (HL-60, MOLM13, and K562), provided by Dr. Toshiya Inaba (Hiroshima University, Hiroshima, Japan), were grown in RPMI-1640 medium containing 10% fetal bovine serum supplemented with penicillin and streptomycin. The detailed procedures for construction of the targeting vector and generation of the cKI mice are described in Fig. 2C. CreERT2 + mice (C57BL/6-Gt(ROSA)26Sortm1(cre/Est1)Arte) were purchased from Taconic Biosciences (Hudson, NY, USA). Cre activation was achieved by intraperitoneal administration tamoxifen (Sigma, T5648). The all animal experiments complied with the ARRIVE guidelines and were conducted in accordance with the Guide for the Care and Use of Laboratory Animals of Hiroshima University Animal Research Committee (permission no. A17-84). Cervical dislocation under light anesthesia was performed as a euthanasia method for mice according to the American Veterinary Medical Association (AVMA) Guidelines. Mouse HSPCs (LSK cells) were stained with antibodies listed in Supplementary Table. 4 and sorted with a FACSAria II (BD Biosciences). LSK cells were grown in S-clone SF-O3 serum-free medium (Iwai North America Inc.) containing 1% BSA supplemented with cytokines (100 ng/mL mouse SCF, 100 ng/mL mouse TPO, 100 ng/mL mouse FLT3L and 50 ng/mL mouse IL-6) (Pepro Tec). CB derived human CD34+ stem/progenitor cells (Lonza) were cultured in StemSpan (Stem Cell Technologies) with 50 ng/mL human SCF (PeproTech), 50 ng/mL human TPO (PeproTech), and 50 ng/mL human Flt3-ligand (PeproTech). The cells were maintained in a humidified, 5% CO2 atmosphere at 37 ℃. Cultured CD34+ cells were stained with antibodies listed in Supplementary Table. 4 and sorted as described above.
Radiation treatment
Cells were irradiated with X-rays using a CP-160 (Acrobio). Total radiation dose was as follows: 5 Gy in human leukemia cell lines and 2 Gy in LSK cells from Ptip+/+ and PtipΔ/Δ mice. After IR, cells were incubated for the indicated time periods at 37 ℃.
Immunofluorescent staining
Cells were plated on poly-L-lysine-coated glass slides (Matsunami Glass), fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100 and treated with Protein block serum-free (Dako) to block non-specific binding. Subsequently, the cells were stained with anti-H3K4me3 and anti-γH2AX antibodies (Supplementary Table. 4). After washing with PBS, the cells were stained with Alexa Fluor 488- or 555-conjugated anti-rabbit and mouse IgG antibodies (Invitrogen). The nuclei were counterstained with Hoechst 33342 (Dojindo Laboratories). Images were taken with an FV-1000 confocal microscope (Olympus) and the fluorescence intensity of individual cells was computationally quantified using ImageJ64 software and were normalized against the mean fluorescence intensity of each control cell type (0 min after IR), as the relative fluorescence intensity34.
Lentiviral vector transduction
To produce lentivirus containing supernatant, pLV-H1-shRNA-EF1α-GFP-Bsd (Biosettia), EF1α-dCas9-VP64-EGFP, Lenti-gRNA-EF1α-BleoR (kindly provided from Dr. Hoshii, Chiba university), Gag-Pol, Rev, and VSV-G expression vectors (Addgene) were transduced into Lenti-X 293 T cells (Takara Bio Inc.) with Lipofectamine 2000 (Invitrogen) and concentrated with Lenti-X concentrator (Takara Bio Inc.). In knockdown experiments, HL-60 cells were infected with lentivirus containing supernatant carrying shCtrl, shPTIP (#1 and #2) and shPRDM1 (#1 and #2). The cell lines were established and expanded before radiation treatment. In dCas9 experiments, HL-60 cells were infected with lentiviral supernatant containing dCas9-VP64 and gRNA (#1 to #4). The shRNA and gRNA sequences are listed in Supplementary Table. 5.
Quantitative real-time PCR
Total cellular RNA was isolated with TRIZOL reagent (Invitrogen) and reverse-transcribed with a SuperScript VILO master mix kit (Invitrogen) according to the manufacturer’s protocol. Quantitative real-time PCR was performed with power SYBR Green PCR Master Mix (Thermo Fisher Scientific) on a StepOnePlus Real-Time PCR system (Applied Biosystems). The primer sequences are listed in Supplementary Table. 5. All data are presented relative to GAPDH.
Histone extraction and western blotting
Cells were resuspended in pH 6.5 lysis buffer (10 mM Tris- HCl at pH 8, 50 mM sodium bisulfite, 1% Triton X-100, 10 mM MgCl2, 8.6% sucrose, 10 mM sodium butyrate). After the supernatant was discarded, the pellet was washed with pH 7.4 wash buffer (10 mM Tris, 13 mM EDTA). After the supernatant was discarded, the pellet was dissolved in 0.4 M H2SO4 for 1 h on ice. The supernatant was collected into a new tube including 80% of acetone and incubated overnight at − 20 ℃. After the supernatant was discarded, the pellets were air-dried and resuspended in water. Histone concentration was assayed using Bradford assay. Three micrograms of total histone extract were loaded into a 15% SDS-PAGE gel. Western blotting was performed as previously described35.
RNA-seq and analysis
Total RNA from duplicate samples (shPTIP #1 and #2) was extracted with RNeasy mini kit (QIAGEN) and RNA-Seq libraries were prepared with the TruSeq Stranded mRNA Library Prep Kit (Illumina). Transcriptome analysis was performed using a next-generation sequencer (HiSeq 2500; Illumina) according to the manufacturer’s instructions. The generated sequence tags (more than 2.0 × 107 reads for each sample) were mapped onto the human genomic sequence (UCSC Genome Browser, version hg19) with the sequence alignment program Tophat. Transcriptomes were compared with FPKM values and analyzed with Gene Set Enrichment Analysis (GSEA_4.3.3) software (Broad Institute, https://www.gsea-msigdb.org/gsea/index.jsp). Gene sets with a false discovery rate (FDR) q-value of < 0.25 were considered statistically significant.
BrdU incorporation analysis
BrdU incorporation was analyzed with a BrdU Flow Kit (BD Biosciences). Pre-cultured HL-60 cells 60 min after IR were treated with BrdU (10 μM) into cultured medium 240 min before the analysis. Cells were fixed, permeabilized and stained with an anti-BrdU antibody conjugated APC (Supplementary Table. 4). BrdU positive cells were measured with a FACSCanto II (BD Biosciences). Data were analyzed with FlowJo v10.10 software (BD Biosciences).
ChIP-qPCR analysis
Chromatin was enzymatically shred and immunoprecipitated with the ChIP-IT Express Enzymatic Kit (Active Motif) according to the manufacturer’s instructions. Antibodies are listed in Supplementary Table. 4. Subsequently, the chromatin was cleaned up with a QIAquick PCR purification kit (QIAGEN). Primer sequences are listed in Supplementary Table. 5.
CUT&RUN assay and analysis
Anti-H3K4me3 and anti-IgG antibodies, listed in Supplementary Table. 4, were used for CUT&RUN performed with the CUT&RUN assay kit (Cell Signaling), according to the manufacturer’s protocol. Libraries were generated using the NEBNext Ultra II DNA Library Prep Kit for Illumina (NEB, E7370L) and sequenced on an Illumina NovaSeq 6000. Paired-end fastq files were processed using the ENCODE Transcription Factor and Histone ChIP-Seq pipeline (https://github.com/ENCODE-DCC/chip-seq-pipeline2). Reads were trimmed using cutadapt v2.5. Reads were aligned to the hg19 genome using Bowtie2 v2.3.4.3, and SAMtools v1.9 was used to convert the output file to BAM format. Duplicates were removed using Picard Tools v2.20.7. Peak calling was performed with MACS2 v2.2.4, and the peaks compared with each IgG sample for subsequent analysis. Narrow peaks (1142–10293 bp, peak score: > 3000) were detected as H3K4me3 enriched peaks, listed in Supplementary Table. 1. Bigwig files were normalized by a scaling factor using Deeptools v3.3.1and bamCoverage tools and visualized in the UCSC genome browser (https://genome.ucsc.edu). The scaling factors were calculated by determining the number of spike-in DNA reads and normalized to total sequencing depth. Homer v4.11 was used for peak annotation and motif analysis. Bedtools v2.29.0 intersect was used to determine overlapping peaks (with at least 1 bp overlap) and assign target genes.
Statistical analysis
The statistical significance of differences between two samples was assessed by two-tailed Student’s t test. When multiple samples were compared, the statistical significance of differences was assessed using one-way ANOVA followed by Dunnett’s test. All statistical tests were performed with Prism software version 6 (GraphPad). All data were obtained from at least two independent experimental replicates. In quantitative real-time PCR, SDs were calculated with technical triplicates, thus statistical significance was not shown.
Data availability
The datasets generated and analyzed during the current study are available in the DNA Data Bank of Japan (DDBJ) repository (https://www.ddbj.nig.ac.jp/) (RNA-seq; DRA010427 and CUT&RUN; DRA016981 (rep1) and DRA018596 (rep2)).
References
Groelly, F. J., Fawkes, M., Dagg, R. A., Blackford, A. N. & Tarsounas, M. Targeting DNA damage response pathways in cancer. Nat. Rev. Cancer 23(2), 78–94 (2023).
Shaltiel, I. A., Krenning, L., Bruinsma, W. & Medema, R. H. The same, only different - DNA damage checkpoints and their reversal throughout the cell cycle. J. Cell Sci. 128(4), 607–620 (2015).
Clay, D. E. & Fox, D. T. DNA damage responses during the cell cycle: Insights from model organisms and beyond. Genes (Basel) 12(12), 1882 (2021).
Zhou, F. X. et al. Prolonged cell cycle arrest in response to DNA damage in yeast requires the maintenance of DNA damage signaling and the spindle assembly checkpoint. eLife https://doi.org/10.7554/eLife.94334.1 (2024).
Mehta, A. & Haber, J. E. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 6(9), a016428 (2014).
Ceccaldi, R., Rondinelli, B. & D’Andrea, A. D. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 26(1), 52–64 (2016).
Hustedt, N. & Durocher, D. The control of DNA repair by the cell cycle. Nat. Cell Biol. 19(1), 1–9 (2016).
Wang, J. et al. PTIP associates with Artemis to dictate DNA repair pathway choice. Genes Dev. 28(24), 2693–2698 (2014).
Wu, J., Prindle, M. J., Dressler, G. R. & Yu, X. PTIP regulates 53BP1 and SMC1 at the DNA damage sites. J. Biol. Chem. 284(27), 18078–18084 (2009).
Jowsey, P. A., Doherty, A. J. & Rouse, J. Human PTIP facilitates ATM-mediated activation of p53 and promotes cellular resistance to ionizing radiation. J. Biol. Chem. 279(53), 55562–55569 (2004).
Munoz, I. M., Jowsey, P. A., Toth, R. & Rouse, J. Phospho-epitope binding by the BRCT domains of hPTIP controls multiple aspects of the cellular response to DNA damage. Nucleic Acids Res. 35(16), 5312–5322 (2007).
Callen, E. et al. 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell 153(6), 1266–1280 (2013).
Cho, Y. W. et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J. Biol. Chem. 282(28), 20395–20406 (2007).
Patel, S. R., Kim, D., Levitan, I. & Dressler, G. R. The BRCT-domain containing protein PTIP links PAX2 to a histone H3, lysine 4 methyltransferase complex. Dev. Cell 13(4), 580–592 (2007).
Cho, Y. W. et al. Histone methylation regulator PTIP is required for PPARgamma and C/EBPalpha expression and adipogenesis. Cell Metab. 10(1), 27–39 (2009).
Callen, E. et al. The DNA damage- and transcription-associated protein paxip1 controls thymocyte development and emigration. Immunity 37(6), 971–985 (2012).
Daniel, J. A. et al. PTIP promotes chromatin changes critical for immunoglobulin class switch recombination. Science 329(5994), 917–923 (2010).
Schwab, K. R., Patel, S. R. & Dressler, G. R. Role of PTIP in class switch recombination and long-range chromatin interactions at the immunoglobulin heavy chain locus. Mol. Cell. Biol. 31(7), 1503–1511 (2011).
Starnes, L. M. et al. A PTIP-PA1 subcomplex promotes transcription for IgH class switching independently from the associated MLL3/MLL4 methyltransferase complex. Genes Dev. 30(2), 149–163 (2016).
Li, X. et al. Histone demethylase KDM5B is a key regulator of genome stability. Proc. Natl. Acad. Sci. U. S. A. 111(19), 7096–7101 (2014).
Bayo, J. et al. Jumonji inhibitors overcome radioresistance in cancer through changes in H3K4 methylation at double-strand breaks. Cell Rep. 25(4), 1040-1050.e5 (2018).
Pai, C. C. et al. A histone H3K36 chromatin switch coordinates DNA double-strand break repair pathway choice. Nat. Commun. 5, 4091 (2014).
Zhang, Y. et al. Histone H3K27 methylation modulates the dynamics of FANCD2 on chromatin to facilitate NHEJ and genome stability. J. Cell Sci. 131(12), jcs215525 (2018).
Kakarougkas, A. et al. Requirement for PBAF in transcriptional repression and repair at DNA breaks in actively transcribed regions of chromatin. Mol. Cell. 55(5), 723–732 (2014).
Rashi-Elkeles, S. et al. Parallel profiling of the transcriptome, cistrome, and epigenome in the cellular response to ionizing radiation. Sci. Signal. 7(325), rs3 (2014).
Esposito, M. T. & So, C. W. DNA damage accumulation and repair defects in acute myeloid leukemia: Implications for pathogenesis, disease progression, and chemotherapy resistance. Chromosoma 123(6), 545–561 (2014).
Capozzo, I., Iannelli, F., Francia, S. & d’Adda di Fagagna, F. Express or repress? The transcriptional dilemma of damaged chromatin. FEBS J. 284(14), 2133–2147 (2017).
Wan, Z., Lu, Y., Rui, L., Yu, X. & Li, Z. PRDM1 overexpression induce G0/G1 arrest in DF-1 cell line. Gene 592(1), 119–127 (2016).
Liu, C. et al. PRDM1 silences stem cell-related genes and inhibits proliferation of human colon tumor organoids. Proc. Natl. Acad. Sci. U. S. A. 115(22), E5066–E5075 (2018).
Bracken, A. P., Ciro, M., Cocito, A. & Helin, K. E2F target genes: Unraveling the biology. Trends Biochem. Sci. 29(8), 409–417 (2004).
Ayrapetov, M. K., Gursoy-Yuzugullu, O., Xu, C., Xu, Y. & Price, B. D. DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin. Proc. Natl. Acad. Sci. U. S. A. 111(25), 9169–9174 (2014).
Gong, F., Clouaire, T., Aguirrebengoa, M., Legube, G. & Miller, K. M. Histone demethylase KDM5A regulates the ZMYND8-NuRD chromatin remodeler to promote DNA repair. J. Cell Biol. 216(7), 1959–1974 (2017).
Fortuny, A. & Polo, S. E. The response to DNA damage in heterochromatin domains. Chromosoma 127(3), 291–300 (2018).
Seita, J. et al. Lnk negatively regulates self-renewal of hematopoietic stem cells by modifying thrombopoietin-mediated signal transduction. Proc. Natl. Acad. Sci. U. S. A. 104(7), 2349–2354 (2007).
Nakata, Y. et al. Acquired expression of CblQ367P in mice induces dysplastic myelopoiesis mimicking chronic myelomonocytic leukemia. Blood 129(15), 2148–2160 (2017).
Ikeda, K. et al. Maintenance of the functional integrity of mouse hematopoiesis by EED and promotion of leukemogenesis by EED haploinsufficiency. Sci. Rep. 6, 29454 (2016).
Sera, Y. et al. UTX maintains the functional integrity of the murine hematopoietic system by globally regulating aging-associated genes. Blood 137(7), 908–922 (2021).
Panchenko, T. et al. Histone H3.3 phosphorylation amplifies stimulation-induced transcription. Nature 583(7818), 852–857 (2020).
Leiendecker, L. et al. LSD1 inhibition induces differentiation and cell death in Merkel cell carcinoma. EMBO Mol. Med. 12(11), e12525 (2020).
Guo, P. et al. Histone variant H3.3 maintains adult haematopoietic stem cell homeostasis by enforcing chromatin adaptability. Nat. Cell Biol. 24(1), 99–111 (2022).
Lee, J. E. et al. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. Elife 2, e01503 (2013).
Callen, E. et al. The DNA damage- and transcription-associated protein paxip1 controls thymocyte development and emigration. Immunity 37(6), 971–985 (2012).
Acknowledgements
We thank Yuki Sakai, Sawako Ogata, and Megumi Nakamura for animal care, genotyping, and molecular experiments as well as Dr. Yoshinori Ohno for technical assistance. We also thank Dr. Junji Takeda (Osaka University, Japan) and RIKEN BRC for providing us with KY1.1 ES cells and B6.Cg-Tg(CAG-Cre)CZ-MO2Osb mice (RBRC01828), respectively. This work was, in part, supported by a JSPS KAKENHI (18K16087), the Kato Memorial Bioscience Foundation and AMED-CREST (JP24gm1310006).
Funding
JSPS KAKENHI,18K16087,Kato Memorial Bioscience Foundation,AMED-CREST,JP24gm1310006.
Author information
Authors and Affiliations
Contributions
Y.N. and H.H. designed the study. H.H., N.Y., and M. K. constructed the targeting vectors and generated genetically engineered mice. Y.N. and A.K. performed RNA-seq analysis. Y.N. and K.K. performed immunofluorescent staining and flow cytometric analysis. Y.N., S.N., and Y.S. performed Cut &Run, ChIP-qPCR and dCas9 assays. Y.N., T.U., I.M., O.K., and H.H. wrote and revised the manuscript. All authors reviewed and agreed on the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Nakata, Y., Nagasawa, S., Sera, Y. et al. PTIP epigenetically regulates DNA damage-induced cell cycle arrest by upregulating PRDM1. Sci Rep 14, 17987 (2024). https://doi.org/10.1038/s41598-024-68295-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-68295-w
- Springer Nature Limited