Abstract
Liquid biopsy has recently emerged as an important tool in clinical practice particularly for lung cancer patients. We retrospectively evaluated cell-free DNA analyses performed at our Institution by next generation sequencing methodology detecting the major classes of genetic alterations. Starting from the graphical representation of chromosomal alterations provided by the analysis software, we developed a support vector machine classifier to automatically classify chromosomal profiles as stable (SCP) or unstable (UCP). High concordance was found between our binary classification and tumor fraction evaluation performed using shallow whole genome sequencing. Among clinical features, UCP patients were more likely to have ≥ 3 metastatic sites and liver metastases. Longitudinal assessment of chromosomal profiles in 33 patients with lung cancer receiving immune checkpoint inhibitors (ICIs) showed that only patients that experienced early death or hyperprogressive disease retained or acquired an UCP within 3 weeks from the beginning of ICIs. UCP was not observed following ICIs among patients that experienced progressive disease or clinical benefit. In conclusion, our binary classification, applied to whole copy number alteration profiles, could be useful for clinical risk stratification during systemic treatment for non-small cell lung cancer patients.
Similar content being viewed by others
Introduction
Current international guidelines recommend daily routine molecular testing using next-generation sequencing (NGS) for actionable genetic alterations in advanced non-small cell lung cancer (NSCLC)1. When tissue biopsy is not sufficient or adequate for molecular characterization, liquid biopsy has been proposed as a tool to increase the availability of molecular characterization in clinical practice. Analyzing cell-free DNA (cfDNA) in plasma potentially provides a minimally invasive approach to diagnose, characterize, monitor the disease and shed light on tumor heterogeneity in cancer patients2,3,4. The detection of targetable genetic alterations at baseline and genetic modifications associated with acquired resistance to targeted agents are currently the most important applications of liquid biopsy. On the other side, cfDNA analysis provides additional information whose potential usefulness for cancer management is under evaluation.
Among these, cfDNA concentration has emerged as a potential prognostic marker in different tumor types5,6,7. In addition, cfDNA concentration emerged as a predictive marker of therapy response in specific contexts, such as locally advanced head and neck cancer, gastric and pancreatic cancer treated with chemotherapy8,9,10. Currently, one of the most promising applications is related to potential predictive value for patients treated with immunotherapy. Although clinical role of immune checkpoint inhibitors (ICIs) is undoubtable in several types of cancers, clinical benefit is highly heterogenous and the identification of predictive biomarkers represents a crucial issue11,12,13. In our previous study in NSCLC patients, longitudinal assessment of cfDNA concentration at baseline and during therapy showed a dramatic increase in cfDNA concentration between baseline and after 3–4 weeks since the start of ICIs in patients experiencing death within 12 weeks since the start of ICIs14.
An additional promising biomarker in cfDNA analysis is based on the definition of the fraction of tumor-derived DNA (tumor fraction, TF), that corresponds to the fraction of cfDNA shed from the tumors (ctDNA). Comprehensive Genomic profiling (CGP) applied to cfDNA analyses permits to define TF by considering aneuploidy or the highest variant allele fraction, excluding germline mutations and specific clonal hematopoiesis (CH)-associated alterations, whenever a tumor is characterized by lack of copy number alterations (CNAs)15. TF varies according to tumor type and during treatment and it is correlated to the number of oncogenic variants and to the level of copy number alterations15. Shallow whole genome sequencing (sWGS) has also been used as a method to estimate TF in cfDNA and depicts the CNAs profile16,17. Changes in cfDNA as detected by sWGS resulted a potential tool to evaluate clinical efficacy of ICIs18,19.
Here, we genetically characterized the cfDNA of a large cohort of NSCLC patients using a commercial assay and we show that, besides detecting somatic alterations in clinical setting, it is possible to extract additional information from chromosomal profiles. We propose a machine learning (ML) approach that allows a binary classification of samples, as stable or unstable, based on chromosomal alteration patterns, and we explore the potential clinical impact of this classification.
Results
A support vector machine (SVM) classifier to predict chromosomal instability in cfDNA samples
AVENIO ctDNA Expanded kit is a capture-based NGS assay covering 77 cancer-associated genes used to detect four types of genetic alterations, including single nucleotide variants (SNVs), insertions/deletions (INDELs), selected CNAs and gene fusions in cfDNA samples. In addition to produce a report including metrics, filtered and unfiltered variants, the software generates a graphical representation of chromosomal alterations detected in cfDNA, which is generally viewed by the operator but it is not further used in downstream data exploitation. When analyzing cfDNA samples from NSCLC patients sent to our laboratory from referral oncologists for diagnostic purposes, we noticed two grossly divergent patterns in the CNA profiles that we defined as SCP or UCP (Fig. 1a,b). The SCP pattern shown in Fig. 1a is similar to that observed in healthy subjects (n = 7, Fig. 1c).
Chromosomal profiles in cfDNA samples analyzed by AVENIO ctDNA Expanded Kit. The image shows two examples of cfDNA samples with a stable pattern (SCP, panel a) and two examples of cfDNA samples with aneuploidy (UCP, panel b), as visually classified by two independent experienced researchers. The panel (c) shows two examples of cfDNA samples from healthy controls (Ctrl). Samples in panels (a) and (b) are from patients, whose plasma was analyzed by NGS with AVENIO ctDNA Expanded kit at diagnosis to assess possible actionable mutations. For each plot, the x-axis represents the loci targeted by the AVENIO Expanded kit, while the y-axis represents the log2 copy ratio observed at these loci. SCP stable chromosomal profile, UCP unstable chromosomal profile, Ctrl healthy donor.
We thus decided to implement an SVM classifier to automatically classify CNA profiles as SCP or UCP, beyond operators’ experience. The first step was the definition of the features to be considered in the classifier. An alteration (“occurrence of instability”) in the CNA profile was defined each time we found a DNA segment of any size with absolute value of the log2 copy ratio exceeding a fixed cut-off. Two different cut-off values on log2 copy ratio were examined: 0.1 and 0.2. Once the cut-off was defined, three features were considered as covariates in the SVM classifier: (1) number of altered segments (Segments), (2) total length of altered regions (Size) and (3) number of affected chromosomes (Chromosomes). In order to classify patients’ samples as SCP or UCP based on AVENIO CNA profiles, we considered the segmented log2 ratios (.cns) files provided by the CNVkit software20 and computed the three features described above: Segments, Size, Chromosomes.
A linear SVM classifier was trained on the 117 samples belonging to the training set, using a repeated tenfold cross validation procedure. Four models were evaluated: the 3-feature classifier (3f) and the three 2-feature classifiers (2f). Details of the best model for each classifier are reported in Table 1 and Supplementary Table 1, for the log2 copy ratio cut-off of 0.1 and 0.2, respectively. Defining of the features based on the 0.1 cut-off yielded higher accuracy both for the 3-feature model and for the three 2-feature models, so we selected this cut-off.
The performance of the four models was assessed on the 60 samples of the test set. As shown in Table 2, all of them performed very well, with overall sensitivity > 0.90 and balanced accuracy > 0.94, regardless of the model. On our dataset, the two-feature classifiers with covariates Segments and Size or Size and Chromosomes performed as well as the three-feature classifier.
For the performance of the four models, the choice to define the features based on a cut-off of 0.1 on the log2 copy ratio was relevant. In fact, Supplementary Table 2 shows that with a cut-off of 0.2 on the log2 copy ratio the number of false negatives would significantly increase, leading to overall sensitivity (min–max: 0.5455–0.7273) and balanced accuracy (min–max: 0.7727–0.8534), both decreased compared to those obtained with the 0.1 cut-off.
To select the final classifier among the three best performers we used the principle of parsimony (2f) and lower correlation between the selected covariates (Size and Chromosomes), Supplementary Fig. 1.
We tested the overall agreement between the 2f Size and Chromosome binary classification and that performed by two independent professionals through visual inspection of CNA profiles. As shown in Supplementary Fig. 2, out of 177 samples evaluated, there were only 5 discordant samples, which were classified as unstable by the human operators and stable by the classifier (Kappa: 0.90), indicating a very high degree of alignment between the two evaluations.
In conclusion, the 2f Size and Chromosome classifier was selected as the best model to substitute experienced researchers in the binary classification of AVENIO CNA profiles as SCP or UCP.
Binary classification of CNA profiles correlates with cfDNA concentration and TF
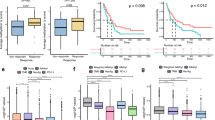
The 2f Size and Chromosome classifier claimed unstable profiles in 28 out of 177 samples (15.8%). When comparing the predicted binary classification with commonly used liquid biopsy parameters, we noticed that cfDNA concentration in plasma was significantly higher in UCP with respect to SCP samples. The median cfDNA concentration of UCP and SCP samples was, respectively, 50.6 ng/ml and 11.2 ng/ml (p < 0.001; Supplementary Table 3). In addition, UCP samples had significantly higher number of tumor-associated variants detected by NGS (p < 0.001; Supplementary Table 3).
In order to understand whether UCP could correlate with a higher tumor fraction in cfDNA, we used sWGS in 12 samples from individual NSCLC patients previously analyzed with the AVENIO ctDNA Expanded kit, according to clinical practice. Among these patients, 4 were classified as SCP and 8 as UCP by the proposed classifier. Data demonstrated a high concordance between the two methods as shown by the representative examples displayed in Fig. 2. Mean TF was 3.6% and 36.6% in SCP and UCP samples, respectively. In particular, all samples defined as UCP presented a TF value > 3% and only 2 samples with a TF value close to the threshold (#43_21 and #54_21) were defined as SCP (Supplementary Table 4). Notably, NGS panel in clinical practice did not detect any tumor-associated genetic variants above the threshold of VAF 0.5% in two cases (#15_20 and #45_20) later classified as UCP and showing an elevated TF (40.7% and 17%, respectively; Supplementary Table 4).
Concordance between sWGS analysis and binary SCP/UCP classification of chromosomal profiles. SCP and UCP representative cases analyzed with AVENIO expanded panel and the binary classifier are shown on the left panel. sWGS analysis and tumor fraction detection were performed in parallel as shown on the right panel. SCP stable chromosomal profile, UCP unstable chromosomal profile.
Association between clinical/pathological features and binary classification in advanced NSCLC
To investigate potential association between clinical/pathological features and the binary classification with the selected SVM classifier, we conducted an analysis involving 84 patients, diagnosed with advanced non-small cell lung cancer, enrolled in our Institute and undergoing NGS analysis in plasma according to clinical practice.
The characteristics of the patients’ cohort and their association with binary classification are summarized in Table 3.
We observed that the presence of three or more metastatic sites and the occurrence of liver metastasis were significantly associated with UCP. On the other hand, it is important to note that no statistically significant association was found between the SCP/UCP binary classification and other traditional prognostic and predictive factors, such as smoking history, PD-L1 status, and the presence of known druggable alterations.
Longitudinal assessment of chromosomal profiles in advanced NSCLC patients treated with immunotherapy
For 33 patients enrolled in MAGIC 1 study and treated with ICIs14 we longitudinally monitored plasma chromosomal profiles during treatment, at baseline (T1) and following one cycle of ICI (T2).
Six cases were UCP and 27 were SCP at T1. In this set of patients, no significant association was found between the UCP/SCP classification and clinical features such as smoking, performance status, histology and positivity to PD-L1 expression. In addition, no association was found with tumor burden, in terms of the number of metastatic sites or the presence of bone and/or liver metastases.
Our binary classification, determined at both T1 and T2, was investigated for association with clinical outcome (Fig. 3). Among 33 enrolled cases, 5 patients experienced progression matching radiological criteria for hyperprogressive disease (HPD), 12 experienced early death (ED), 8 experienced progressive disease (PD) without matching HPD criteria and 8 showed clinical benefit (CB). One patient (M#43) met radiological HPD criteria and experienced ED. The 2f Size and Chromosome classifier identified 9 UCP patients, showing the unstable profile at least one time point (T1, T2 or both). Importantly, 6 out of 9 patients presented with UCP at both T1 and T2 and were included in HPD (n = 1) and ED (n = 5) groups. Among the remaining UCP patients, 2 were UCP at T1 and became SCP after therapy. Notably, these 2 patients were both included in the CB group. Finally, the last patient resulted SCP at T1 and presented UCP after immunotherapy and, importantly, this patient was included in the HPD group.
Association between the SCP/UCP binary classification and clinical outcome. SCP/ UCP classification was performed in 33 patients enrolled in the MAGIC-1 study both at baseline (T1) and following one cycle of ICIs (T2). For clinical outcome, patients were divided in 2 groups according the presence or absence of potential detrimental effects (ED + HPD and PD + CB, respectively). HPD hyperprogressive disease, ED early death, PD progressive disease, CB clinical benefit, SCP stable chromosomal profile, UCP unstable chromosomal profile.
Overall, 7 out of 9 patients, experiencing either HPD or ED, had UCP after the beginning of ICIs treatment, while, amongst those not experiencing potential detrimental effects, none of the patients presented UCP after ICIs (Fig. 3).
In Fig. 4, representative examples of UCP patients that experienced CB and ED are shown. Amongst patients with CB, samples M#185 and M#251 were UCP at T1 and switched to SCP at T2 (top panel). In contrast, M#191 and M#301 patients, that experienced ED, presented with UCP at diagnosis (T1), showed no change at T2 and poor response to immunotherapy (bottom panel).
Representative analyses of dynamic assessment of CNAs and response to immunotherapy. Representative plots of CNAs generated using AVENIO ctDNA Expanded Kit for patients undergoing immunotherapy treatment. Analyses were performed at baseline (T1) and following 1 cycle of ICI (T2); T3 represents CT scan re-evaluation. In the upper panel, plots of two patients that belong to the CB group are shown (M#185 and M#251). In the lower panel, two patients that experienced ED are included (M#191 and M#301). ED early death, CB clinical benefit, SCP stable chromosomal profile, UCP unstable chromosomal profile.
Discussion
Liquid biopsy is an innovative tool whose exploitation both in translational research and in clinical practice is rapidly increasing3,4,21. Most promising applications in the next features are the detection and monitoring of minimal residual disease in early-stage cancer patients and the dynamic evaluation of changes induced by systemic treatment in advanced diseases22. In particular, our group focused on the study of longitudinal liquid biopsy as potential predictive marker for advanced NSCLC patients treated with ICIs14.
Even though the idea of monitoring disease and anticipating treatment long-term efficacy by studying tumor-associated alterations in blood is fascinating and widely accepted among scientific community, the practical application is highly challenging mainly due to technical issues. Among them, we would like to highlight the lack of standardized methods to quantitatively define tumor burden in plasma and the difficulties to perform wide genetic characterization in clinical practice.
In the current manuscript, we propose a ML approach to extract additional information from cfDNA analysis of a relatively small NGS liquid biopsy assay used for clinical practice genetic characterization and at no additional cost. For this purpose, we retrospectively evaluated the plasma NGS analysis of 177 samples performed at our Institution by using the AVENIO Expanded Kit, a panel of 77 genes, able to detect the main classes of genetic alterations. Our belief was that, alongside the specific information on the individual alterations, the whole alteration pattern that the analysis software represented only graphically, without further exploitation, could add relevant information about the sample. Starting from the observation, made by our expert researchers, of two grossly divergent chromosomal profiles (SCP and UCP), we wanted to investigate the potential relevancy of this classification in relation to known clinical/pathological parameters and as predictive marker of response to ICIs treatment in NSCLC. The SCP/UCP classification, performed by visual inspection of CNA profile graphs by two independent professionals, was used as a target to train an ML model and automate the classification procedure. Available samples were thus split into a training set and a test set. To extract the entire AVENIO CNA profiles, we considered the segmented log2 ratios (.cns) files provided by the AVENIO CNVkit software. On these data, we computed the three features (Segments, Size, Chromosomes) described in the "Methods" section. Four linear SVM classifiers, one with three features as covariates and the others based on the two-feature combinations, were developed and their parameters optimized. It is worth noting that the choice of the 0.1 cut-off on the log2 copy ratio used to define an “occurrence of instability”, and consequently to calculate the three features (Segments, Size and Chromosomes), had a major impact on the performance of the models. Based on the double criterion of parsimony and lower correlation between covariates, the 2f Size and Chromosome classifier was selected as the best model to substitute experienced researchers in the binary classification of AVENIO CNA profiles as SCP or UCP.
Notably, UCP samples strongly correlated with a positive tumor fraction as determined by shallow whole genome sequencing, a validated method to quantify TF and to detect CNAs widely used in liquid biopsy. CNAs are distinctive traits of tumor cells23, differently from somatic nucleotide variants that can occur also in healthy individuals, as for examples for SNV associated with hereditary syndromes (germline mutations) or clonal hematopoiesis24,25. For this reason, exploiting a relatively cheap technique as sWGS is being widely used to quantify the fraction of ctDNA in liquid biopsy samples. This approach is particularly suitable to refine the interpretation of samples that were classified as ctDNA-negative by mutation-based strategies and can provide guidance to properly select downstream analyses26. This approach is also widely used in early cancer detection27,28 and as marker of tumor progression after systemic treatment18. Generally, threshold for TF determination using sWGS is around 3% and samples having ctDNA fraction below this cut-off might not be informative and require more sensitive approaches17. Therefore, using GCP analysis, that allows integration of CNAs with variant allele fraction and canonical alteration, it is possible to lower the threshold, down to 1%29. The characterization of TF can improve the reliability of a liquid biopsy test in particular when applied to clinical management of patients with advanced disease. In fact, recent guidelines on liquid biopsy underline the importance of specifying whether results coming from cfDNA analysis are informative or not. In particular, the detection of TF ≥ 1% in patients without genetic alterations found in plasma is suggested as a tool to avoid tissue re-biopsy29.
In this context, it is worth mentioning that our binary classification was found to be consistent with another validated method (sWGS), considering that SCP and UCP samples had significantly different TFs (3.6% and 36.6%, respectively). It is also important to underline that, besides some similarities, our approach does not return a quantitative score associated to a limited threshold but a binary output that, combined with the detection of genetic alterations, allows, with a relatively low number of genes, both genetic information useful for clinical practice and the identification of cases with specific biological features. Notably, all the samples defined as UCP were characterized by a TF > 3% and by peculiar clinical/biological features.
In fact, in our series of NSCLC patients recruited and treated at our Institute, UCP was associated with tumor burden and presence of liver metastases, thus suggesting a potential negative prognostic value of profiles classified as unstable. On the other hand, it was neither associated with other clinical prognostic factors nor with commonly used molecular predictive markers, such as PD-L1 and presence of driver alterations (Table 3). This point hints the potential of the new classification to be integrated with other known prognostic and predictive markers in a multivariate statistical model.
Importantly, using data from NSCLC patients treated with immunotherapy (MAGIC-1 clinical study), we highlighted the possible usefulness of our classifier after the start of systemic treatment, when it is likely to have a higher predictive value. Although our results need to be confirmed in a larger cohort, we could appreciate a high correlation between detection of UCP after treatment and patients that experienced either HPD or ED. None of patients not experiencing potential detrimental effects acquired UCP after the first cycle of ICIs (T2). Importantly, classification of chromosomal profile status at different time points can add important information for those patients who do not have mutations at baseline; this is the case of the M#185 sample that presented a switch from UCP to SCP, but no other alterations at T1 to track during therapy. These results suggest the rationale for longitudinal liquid biopsy assessments during treatment that should be implemented in prospective clinical studies14,22,30,31,32,33. In conclusion, our study demonstrates that it is possible to extract additional information from an NGS liquid biopsy gene panel already used for clinical practice genetic characterization and at no additional cost. By considering the whole CNA profiles, through ML techniques, we binary classified chromosomal profiles and showed that the UCP status can be regarded as a novel parameter to be evaluated in liquid biopsy and integrated with other commonly used prognostic/predictive parameters. To be applied routinely, the proposed binary classifier requires further validation in a larger cohort of patients and the development of an easy-to-use tool for researches without specific bioinformatics expertise.
Materials and methods
Patients, plasma sample collection and study design
From January 2020 to March 2021, 100 advanced cancer patients underwent liquid biopsy according to clinical practice and their samples were analyzed by using the ctDNA AVENIO Expanded Kit—a NGS liquid biopsy assay containing a 77 pan-cancer gene panel (Roche Sequencing Solutions, Pleasanton, CA). All the patients signed informed consent to perform plasma NGS analysis.
An additional 77 samples were collected for different research projects and included 8 EGFR-mutated tumors after histological transformation in small-cell lung cancer, enrolled in ESTRA study, and 33 advanced EGFR-ALK-ROS1 wild-type NSCLC patients treated with immunotherapy from January 2017 to August 2019 and enrolled in MAGIC-1 clinical study14.
ESTRA clinical study was approved by the Istituto Oncologico Veneto Ethics Committee (protocol number 2021/13, 25/01/2021). Written informed consent was obtained from the participants or their legal guardians. In this study plasma samples were collected at the time of histological transformation.
MAGIC-1 clinical study was approved by the Istituto Oncologico Veneto Ethics Committee (protocol number 2016/82, 12/12/2016). Written informed consent was obtained from the participants or their legal guardians. In this study, plasma samples were collected at baseline (T1), after 3/4 weeks of treatment (T2), at the CT scan re-evaluation (T3) and at radiological progression (T4).
NGS was performed for all the 177 samples using the AVENIO Expanded Kit for cfDNA analysis.
For the construction of a binary classifier to predict SCP or UCP, all available samples were included, even those collected from the same patients at different time points. Specifically, 117 randomly selected cases were used as the training set for the classifier and the model’s performance in classifying profiles as stable or unstable was assessed on the remaining 60 cases in the test set.
Amongst all the samples analyzed, the correlation between the proposed binary classification and the clinical characteristics was performed only in advanced NSCLC patients enrolled and treated at our Institution (n = 84).
Among patients enrolled in MAGIC-1 clinical study an exploratory evaluation of chromosomal profile modification during treatment was performed.
All methods were performed according to the relevant guidelines and regulations.
Samples, cfDNA extraction and sequencing
For all samples, 20 ml of peripheral blood were collected in two Helix cfDNA Stabilization tubes (Streck Corporate, La Vista, NE, USA) and processed within 24–72 h, as previously described22. Briefly, blood sample was centrifuged at 2000×g for 10 min at 4 °C and the supernatant was subsequently centrifuged at 20,000×g for 10 min. Plasma samples were stored at − 80 °C until the analysis.
Circulating free DNA (cfDNA) was extracted from 2 to 5 ml of plasma using the AVENIO cfDNA Isolation Kit (Roche Sequencing Solutions, Pleasanton, CA), according to manufacturer’s instructions, and eluted into 60 µL of buffer, as previously described34. Sequencing libraries were prepared from 10 to 50 ng cfDNA, using the AVENIO ctDNA Expanded kit (77 genes; Roche Diagnostics, Basilea, CHE), according to the manufacturer’s instructions. Four or eight purified libraries per run were pooled and sequenced on an Illumina NextSeq 500 (Illumina, San Diego, CA, USA), using the 300-cycle NextSeq 500/550 Mid Output v2 kit or the 300-cycle NextSeq High Output kit, respectively, in paired-end mode (2 × 151 cycles).
Targeted sequencing analysis using AVENIO ctDNA Expanded kit
Following sequencing, alignment and gene variant calling were performed using the AVENIO Oncology Analysis Software (Roche Sequencing Solutions, Pleasanton, CA), with default parameter settings for the expanded panel. The analysis software includes three default reports that are automatically generated: a sample metrics report, an initial variant report (unfiltered listing all variants), and a second variant report (Roche default filter) which highlights known somatic mutations and discards known polymorphisms based on annotation databases. The percentage of aligned reads to the human genome that are within the targeted region (unique depth) according to the manufacturer’s instructions should be > 40%. Similarly, the expected median unique depth across bases in the targeted region should be at least 2500×, given 50 ng input cfDNA. All variants were manually inspected and gene variants present in population databases (ExAC, dbSNP, 1000 genomes) were not considered as relevant. Variants were considered reliable with a VAF > 0.5%. To investigate pathogenicity value, the target variants were submitted to the disease-associated databases COSMIC35, VARSOME36 and OncoKB37, and only variants annotated as pathogenic or likely pathogenic were considered. The following 77 genes are included in the AVENIO ctDNA Expanded kit: ABL1, AKT1, AKT2, ALK, APC, AR, ARAF, BRAF, BRCA1, BRCA2, CCND1, CCND2, CCND3, CD274, CDK4, CDK6, CDKN2A, CSF1R, CTNNB1, DDR2, DPYD, EGFR, ERBB2, ESR1, EZH2, FBXW7, FGFR1, FGFR2, FGFR3, FLT1, FLT3, FLT4, GATA3, GNA11, GNAQ, GNAS, IDH1, IDH2, JAK2, JAK3, KDR, KEAP1, KIT, KRAS, MAP2K1, MAP2K2, MET, MLH1, MSH2, MSH6, MTOR, NF2, NFE2L2, NRAS, NTRK1, PDCD1LG2, PDGFRA, PDGFRB, PIK3CA, PIK3R1, PMS2, PTCH1, PTEN, RAF1, RB1, RET, RNF43, ROS1, SMAD4, SMO, STK11, TERT, TP53, TSC1, TSC2, UGT1A1, VHL.
ML approach
A ML approach based on a linear SVM classifier was applied to predict chromosomal instability. Binary classification into SCP and UCP, performed by visual inspection of individual profiles by two independent professionals (S.I. and E.Z.), was used as target. Analysis was performed in the R statistical environment version 4.4.0 (R Foundation for Statistical Computing http://www.r-project.org/). Available samples (n = 177) were randomly partitioned into training (n = 117, 66%) and testing (n = 60, 34%) sets using the function sample.split() in the ‘caTools’ package. Linear SVM models were trained using the ‘caret’ package38. A repeated tenfold cross validation with 3 repeats was used to assess the model performance on the training set and meanwhile to select the best cost parameter, C ∈ {0.01, 0.1, 1, 10, 100}. Pre-processing transformations (centering and scaling) were estimated from the training data and then applied to test data. Type of cross validation (repeated CV) as well as the number of cross validation folds and the number of repeats were specified with the trainControl() function, which was passed to the trControl argument in train() function implemented in the ‘caret’ package. The best classifier was then used to predict the chromosomal instability for the, previously unseen, samples in the test set, using the function predict(). The performance was evaluated in terms of accuracy and area under the receiver operating characteristic (ROC) curve using the ‘ROCR’ package. The ‘ggplot2’ package was used for graphical visualization.
Whole genome libraries preparation and sequencing
sWGS libraries were prepared starting from 10–20 ng of cfDNA using the KAPA Hyper Prep Kit with KAPA Dual-Indexed Adapters for Illumina platforms (Roche Sequencing Solutions, Pleasanton, CA). Briefly, after sequencing adapter ligation for 15 h at 20 °C, DNA libraries were purified by double-sided size selection to selectively capture DNA fragments comprised between 150 and 350 bp. Adapter-ligated libraries were amplified in 11 PCR cycles. Final libraries were diluted to a concentration of 10 nM and pooled in equimolar amount to a final sequencing concentration of 1 pM. Libraries were sequenced using 150-bp paired-end runs on a High output flow cell on a NextSeq 550 platform (Illumina) to average genome-wide fold coverage of 0.5×.
We used the ichorCNA tools package16 to evaluate the fraction of tumor in cfDNA and predict locations of CNAs at the same time. For plasma samples, the tumor fraction (TF) was calculated, and the presence of ctDNA was indicated by setting the cut-off of 0.03 (a sensitivity threshold identified by ichorCNA). Plasma samples that failed quality checks on sWGS analysis (coverage > 0.1× and mean absolute deviation = MAD < 0.150) were excluded from the analysis. Only autosomal chromosomes were taken into account for CNA analysis.
Statistical analyses
To investigate possible associations between clinical characteristics and chromosomal profile clinical data were retrospectively collected from patients’ medical records. The radiological response was assessed using RECIST criteria v 1.139. CB was defined as the lack of progression within six months since the start of systemic treatment. Patients who had at least two computed tomography (CT) scans available prior to the initiation of ICIs treatment were evaluated to assess the presence of HPD. Tumor Growth Rate (TGR) was defined based on established criteria40,41 with PD being classified as HPD when the TGR during ICI treatment exceeded 50% of the TGR measured before ICIs initiation42. ED was defined as death within 12 weeks since the start of systemic treatment.
Statistical tests were performed by Fisher's exact test or Wilcoxon rank sum test, as deemed appropriate. Statistical analysis was conducted using R software version 4.4.0 (R Foundation for Statistical Computing http://www.r-project.org/).
Data availability
Data underlying the SVM classifier development are available in Zenodo repository, https://doi.org/10.5281/zenodo.11366939. Clinical data are available upon request to the corresponding author.
Code availability
Code is available upon request to the corresponding author.
References
Mosele, F. et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. 31, 1491–1505. https://doi.org/10.1016/j.annonc.2020.07.014 (2020).
Nikanjam, M., Kato, S. & Kurzrock, R. Liquid biopsy: Current technology and clinical applications. J. Hematol. Oncol. 15, 131. https://doi.org/10.1186/s13045-022-01351-y (2022).
Rolfo, C. et al. Liquid biopsy for advanced NSCLC: A consensus statement from the international association for the study of lung cancer. J. Thorac. Oncol. 16, 1647–1662. https://doi.org/10.1016/j.jtho.2021.06.017 (2021).
Bonanno, L. et al. Liquid biopsy and non-small cell lung cancer: Are we looking at the tip of the iceberg?. Br. J. Cancer 127, 383–393. https://doi.org/10.1038/s41416-022-01777-8 (2022).
Tissot, C. et al. Circulating free DNA concentration is an independent prognostic biomarker in lung cancer. Eur. Respir. J. 46, 1773–1780. https://doi.org/10.1183/13993003.00676-2015 (2015).
Cheng, J. et al. Circulating free DNA integrity and concentration as independent prognostic markers in metastatic breast cancer. Breast Cancer Res. Treat. 169, 69–82. https://doi.org/10.1007/s10549-018-4666-5 (2018).
Varaljai, R. et al. The predictive and prognostic significance of cell-free DNA concentration in melanoma. J. Eur. Acad. Dermatol. Venereol. 35, 387–395. https://doi.org/10.1111/jdv.16766 (2021).
Koukourakis, M. I. et al. Circulating plasma cell-free DNA (cfDNA) as a predictive biomarker for radiotherapy: Results from a prospective trial in head and neck cancer. Cancer Diagn. Progn. 3, 551–557. https://doi.org/10.21873/cdp.10254 (2023).
Zhong, Y. et al. Plasma cfDNA as a potential biomarker to evaluate the efficacy of chemotherapy in gastric cancer. Cancer Manag. Res. 12, 3099–3106. https://doi.org/10.2147/CMAR.S243320 (2020).
Christenson, E. S. et al. Cell-free DNA predicts prolonged response to multi-agent chemotherapy in pancreatic ductal adenocarcinoma. Cancer Res. Commun. 2, 1418–1425. https://doi.org/10.1158/2767-9764.CRC-22-0343 (2022).
Sankar, K. et al. The role of biomarkers in personalized immunotherapy. Biomark. Res. 10, 32. https://doi.org/10.1186/s40364-022-00378-0 (2022).
Alessi, J. V. et al. Clinicopathologic and genomic factors impacting efficacy of first-line chemoimmunotherapy in advanced NSCLC. J. Thorac. Oncol. 18, 731–743. https://doi.org/10.1016/j.jtho.2023.01.091 (2023).
Otano, I., Ucero, A. C., Zugazagoitia, J. & Paz-Ares, L. At the crossroads of immunotherapy for oncogene-addicted subsets of NSCLC. Nat. Rev. Clin. Oncol. 20, 143–159. https://doi.org/10.1038/s41571-022-00718-x (2023).
Zulato, E. et al. Longitudinal liquid biopsy anticipates hyperprogression and early death in advanced non-small cell lung cancer patients treated with immune checkpoint inhibitors. Br. J. Cancer 127, 2034–2042. https://doi.org/10.1038/s41416-022-01978-1 (2022).
Husain, H. et al. Tumor fraction correlates with detection of actionable variants across > 23,000 circulating tumor DNA samples. JCO Precis. Oncol. 6, e2200261. https://doi.org/10.1200/PO.22.00261 (2022).
Adalsteinsson, V. A. et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat. Commun. 8, 1324. https://doi.org/10.1038/s41467-017-00965-y (2017).
Rickles-Young, M. et al. Assay validation of cell-free DNA shallow whole-genome sequencing to determine tumor fraction in advanced cancers. J. Mol. Diagn. 26, 413–422. https://doi.org/10.1016/j.jmoldx.2024.01.014 (2024).
Carbonell, C. et al. Dynamic changes in circulating tumor DNA assessed by shallow whole-genome sequencing associate with clinical efficacy of checkpoint inhibitors in NSCLC. Mol. Oncol. 17, 779–791. https://doi.org/10.1002/1878-0261.13409 (2023).
Sivapalan, L. et al. Dynamics of sequence and structural cell-free DNA landscapes in small-cell lung cancer. Clin. Cancer Res. 29, 2310–2323. https://doi.org/10.1158/1078-0432.CCR-22-2242 (2023).
Talevich, E., Shain, A. H., Botton, T. & Bastian, B. C. CNVkit: Genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput. Biol. 12, e1004873. https://doi.org/10.1371/journal.pcbi.1004873 (2016).
Ferro, A. et al. The study of primary and acquired resistance to first-line osimertinib to improve the outcome of EGFR-mutated advanced Non-small cell lung cancer patients: The challenge is open for new therapeutic strategies. Crit. Rev. Oncol. Hematol. 196, 104295. https://doi.org/10.1016/j.critrevonc.2024.104295 (2024).
Zulato, E. et al. Early assessment of KRAS mutation in cfDNA correlates with risk of progression and death in advanced non-small-cell lung cancer. Br. J. Cancer 123, 81–91. https://doi.org/10.1038/s41416-020-0833-7 (2020).
Steele, C. D. et al. Signatures of copy number alterations in human cancer. Nature 606, 984–991. https://doi.org/10.1038/s41586-022-04738-6 (2022).
Vasseur, D. et al. Genomic landscape of liquid biopsy mutations in TP53 and DNA damage genes in cancer patients. NPJ Precis. Oncol. 8, 51. https://doi.org/10.1038/s41698-024-00544-7 (2024).
Stout, L. A. et al. Identification of germline cancer predisposition variants during clinical ctDNA testing. Sci. Rep. 11, 13624. https://doi.org/10.1038/s41598-021-93084-0 (2021).
Tsui, D. W. Y. et al. Tumor fraction-guided cell-free DNA profiling in metastatic solid tumor patients. Genome Med. 13, 96. https://doi.org/10.1186/s13073-021-00898-8 (2021).
Mouliere, F. et al. Detection of cell-free DNA fragmentation and copy number alterations in cerebrospinal fluid from glioma patients. EMBO Mol. Med. https://doi.org/10.15252/emmm.201809323 (2018).
Szymanski, J. J. et al. Cell-free DNA ultra-low-pass whole genome sequencing to distinguish malignant peripheral nerve sheath tumor (MPNST) from its benign precursor lesion: A cross-sectional study. PLoS Med. 18, e1003734. https://doi.org/10.1371/journal.pmed.1003734 (2021).
Rolfo, C. D. et al. Measurement of ctDNA tumor fraction identifies informative negative liquid biopsy results and informs value of tissue confirmation. Clin. Cancer Res. https://doi.org/10.1158/1078-0432.CCR-23-3321 (2024).
Anagnostou, V. et al. ctDNA response after pembrolizumab in non-small cell lung cancer: Phase 2 adaptive trial results. Nat. Med. 29, 2559–2569. https://doi.org/10.1038/s41591-023-02598-9 (2023).
Vega, D. M. et al. Changes in circulating tumor DNA reflect clinical benefit across multiple studies of patients with non-small-cell lung cancer treated with immune checkpoint inhibitors. JCO Precis. Oncol. 6, e2100372. https://doi.org/10.1200/PO.21.00372 (2022).
Goldberg, S. B. et al. Early assessment of lung cancer immunotherapy response via circulating tumor DNA. Clin. Cancer Res. 24, 1872–1880. https://doi.org/10.1158/1078-0432.CCR-17-1341 (2018).
Thompson, J. C. et al. Serial monitoring of circulating tumor DNA by next-generation gene sequencing as a biomarker of response and survival in patients with advanced NSCLC receiving pembrolizumab-based therapy. JCO Precis. Oncol. https://doi.org/10.1200/PO.20.00321 (2021).
Zulato, E. et al. Implementation of next generation sequencing-based liquid biopsy for clinical molecular diagnostics in non-small cell lung cancer (NSCLC) Patients. Diagnostics (Basel) https://doi.org/10.3390/diagnostics11081468 (2021).
Tate, J. G. et al. COSMIC: The catalogue of somatic mutations in cancer. Nucleic Acids Res. 47, D941–D947. https://doi.org/10.1093/nar/gky1015 (2019).
Kopanos, C. et al. VarSome: The human genomic variant search engine. Bioinformatics 35, 1978–1980. https://doi.org/10.1093/bioinformatics/bty897 (2019).
Suehnholz, S. P. et al. Quantifying the expanding landscape of clinical actionability for patients with cancer. Cancer Discov. 14, 49–65. https://doi.org/10.1158/2159-8290.CD-23-0467 (2024).
Kuhn, M. Building predictive models in R using the caret package. J. Stat. Softw. 28, 1–26. https://doi.org/10.18637/jss.v028.i05 (2008).
Eisenhauer, E. A. et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur. J. Cancer 45, 228–247. https://doi.org/10.1016/j.ejca.2008.10.026 (2009).
Gomez-Roca, C. et al. Tumour growth rates and RECIST criteria in early drug development. Eur. J. Cancer 47, 2512–2516. https://doi.org/10.1016/j.ejca.2011.06.012 (2011).
Ferte, C. et al. Tumor growth rate is an early indicator of antitumor drug activity in phase I clinical trials. Clin. Cancer Res. 20, 246–252. https://doi.org/10.1158/1078-0432.CCR-13-2098 (2014).
Ferrara, R. et al. Hyperprogressive disease in patients with advanced non-small cell lung cancer treated with PD-1/PD-L1 inhibitors or with single-agent chemotherapy. JAMA Oncol. 4, 1543–1552. https://doi.org/10.1001/jamaoncol.2018.3676 (2018).
Acknowledgements
We would like to thank Marinelli Marilisa for her continuous support with the AVENIO ctDNA Expanded NGS assay.
Funding
This work was funded by IOV intramural research grant 2017–5×1000 (MAGIC-2, to S. Indraccolo and L. Bonanno) and Ricerca corrente 2024 funding from the Italian Ministry of Health.
Author information
Authors and Affiliations
Contributions
Valeria Tosello and Angela Grassi contributed equally to this manuscript. V.T. and E.Z. performed experiments; A.G. and D.R processed NGS experimental data and developed the linear SVM classifier for binary classification; E.Z. and S.I. analyzed graphic representation of aneuploidy; C.D.F and M.P. performed and analyzed sWGS; C.L.B and L.B performed and interpreted clinical analyses; P.D.B. performed statistical analysis; V.T., E.Z., S.I. and L.B. planned and supervised experiments. G.P. and V.G. provided clinical samples. V.T., A.G. and L.B. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
Dominic Rose is affiliated with Roche Diagnostics Deutschland GmbH, which is the company that provided the assay for this study. The other authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Tosello, V., Grassi, A., Rose, D. et al. Binary classification of copy number alteration profiles in liquid biopsy with potential clinical impact in advanced NSCLC. Sci Rep 14, 18545 (2024). https://doi.org/10.1038/s41598-024-68229-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-68229-6
- Springer Nature Limited