Abstract
SLAMF9, a member of the conserved lymphocyte activation molecules family (SLAMF), has been less investigated compared to other SLAMs, especially concerning its implications across various cancer types. In our systematic pan-cancer investigation, we observed elevated SLAMF9 expression in various tumor tissues, which was correlated with reduced patient survival across most malignancies. Correlation analyses further revealed significant associations between SLAMF9 expression and immune cell infiltrates, immune checkpoint inhibitors, tumor mutation load, microsatellite instability, and epithelial–mesenchymal transition (EMT) scores. Cell-based assays demonstrated that SLAMF9 knockdown attenuated the proliferative, motile, and invasive capacities of colorectal cancer (CRC) cells. In a nude mouse xenograft model, suppression of SLAMF9 expression substantially inhibited tumor growth. These findings highlight the potential of SLAMF9 as a prognostic and therapeutic biomarker across tumors, with notable implications for CRC cell proliferation and migration.
Similar content being viewed by others
Introduction
Malignant tumors have emerged as a significant threat to human life and health, with a notable surge in both incidence and mortality rates over recent decades. This alarming trend has been attributed to various factors, including genetic predispositions, exposure to external agents such as chemical, physical and biological carcinogens, and alterations in lifestyle habits1,2. Regrettably, effective treatments for cancer remain elusive. Recent advances in cancer research have highlighted the utility of pan-cancer analysis, particularly in light of the continuous improvements and expansion of public databases such as The Cancer Genome Atlas (TCGA). Through such analyses, researchers can identify novel therapeutic targets by assessing gene expression patterns across diverse cancer types and assessing their associations with clinical prognosis3,4,5.
The signaling lymphocyte activating molecule (SLAM) family comprises receptors unique to hematopoietic cells6. This family can be categorized into two groups based on their cytoplasmic signaling motif: classical and non-classical7. Classical SLAM family members, such as SLAMF1-7, typically possess two to four immunoreceptor tyrosine switch motifs (ITSMs), which facilitate the recruitment of molecules with SH2 domains, including the SLAM-associated protein (SAP) and other phosphatases. These receptors play important roles in various immunological processes, such as NK-T cell formation, NK cell activation, and follicular T helper cell differentiation8,9. SLAMF8 and SLAMF9 are the most frequently studied non-classical SLAM family members10. Unlike traditional SLAMF receptors, they lack signaling elements in their short cytoplasmic tail. These receptors are selectively expressed by various myeloid cells, including neutrophils, dendritic cells (DCs), and macrophages. SLAMF9, a member of the conserved lymphocyte activation molecules family, has been relatively underexplored in malignancy research compared to other SLAM family members. The SLAMF9 gene (also known as CD2F10, SF2001, CD84H) is located outside the SLAM locus7. Specifically, the transcript of SLAMF9 is predominantly present in fringe monocytes and human monocyte-determined DCs. However, studies on SLAMF9 have typically focused on individual or small subsets of cancer types, lacking comprehensive comparisons across different malignancies to delineate their similarities and differences. A thorough examination is urgently needed because this information is crucial for comprehending the role of SLAMF9 in diverse malignancies.
The present study aimed to investigate SLAMF9 expression profiles and their prognostic implications in human malignancies, utilizing the extensive datasets from TCGA. Additionally, the association between SLAMF9 expression and levels of tumor infiltration, tumor mutational burden (TMB), and microsatellite instability (MSI) across various tumor types was explored. Gene set enrichment analysis (GSEA) was conducted to elucidate potential underlying mechanisms. Furthermore, molecular biology verification was performed in colorectal cancer (CRC) to reinforce SLAMF9’s role in neoplasm promotion. Taken together, this study provides valuable insights into the significant role of SLAMF9 in cancers, identifies potential connections between SLAMF9 and tumor-immune interactions, and highlights possible underlying mechanisms.
Materials and methods
Data source and processing
We downloaded pan-cancer sequencing data from various cancer samples in the TCGA database and public SLAMF9 expression data of normal tissue from the Genotype-Tissue Expression (GTEx:https://commonfund.nih.gov/GTEx/) by using UCSC Xena (https://xenabrowser.net/, last accessed February 18, 2024)11. Normal samples from both the TCGA and GTEx databases (http://commonfund.nih.gov/GTEx/) were utilized for comparisons between cancer and normal tissue. Additionally, data on tumor mutation burden (TMB) and MSI were exclusively obtained from the TCGA database. TMB was calculated by tallying the number of insertion or deletion events in repetitive gene sequences, while MSI was assessed by counting the overall mutation occurrences per million base pairs.
SLAMF9 survival analysis
The Kaplan–Meier Plotter database (https://kmplot.com/analysis/, last accessed February 18, 2024) and log-rank test were used to obtain the overall survival (OS) of cases expressing SLAMF9 across various cancers12. Additionally, the PrognoScan database (https://dna0o.bio.kyutech.ac.jp/PrognoScan/index.html, last accessed February 18, 2024) was utilized to investigate associations between SLAMF9 expression levels and survival outcomes, including OS and progression-free intervals (PFIs). The results were visualized using forest plots generated with the R package. Univariate Cox proportional hazards models were applied to evaluate correlations between clinical profiles of specimens and patients, with a significance threshold of P < 0.05 indicating meaningful differences in specific cancers. Hazard ratios (HRs), log-rank P values, and 95% confidence intervals (CIs) were estimated as part of the analysis.
SLAMF9 tumor immune invasion
The TIMER2.0 database (https://timer.cistrome.org/, last accessed February 18, 2024) was utilized to examine SLAMF9 expression and immune cell infiltration13. This database offers six advanced algorithms, including TIMER, xCell, MCP-counter, CIBERSORT, EPIC, and quanTIseq. The ESTIMATE algorithm within the R package was employed to analyze the ratio of immune matrix components in the tumor microenvironment (TME). The results were presented as three scores: ImmuneScore, ESTIMATE, and Stromal Score, which are positively correlated with immune cells, extracellular matrix, and the sum of the two, respectively14. These scores were positively associated with the corresponding component ratio in the TME. Additionally, the relationship between SLAMF9 expression and immune checkpoint markers was analyzed using Spearman’s correlation analysis.
Additionally, we used Gene Expression Profiling Interactive Analysis (GEPIA) (https://gepia.cancer pku.cn/index.html, last accessed February 18, 2024), a web-based tool facilitating in-depth exploration of the TCGA and GTEx databases15 to conduct gene correlation analysis, complementing our comprehensive analysis.
Gene set enrichment analysis
GSEA (https://www.gsea-msigdb.org/gsea/index.jsp, last accessed February 18, 2024) was used to compare biological signaling pathways between the SLAMF9 high-expression group and the low-expression group16,17. Visualization of enrichment map results was achieved using Bioconductor (http://bioconductor.org/) and R software (http://www.r-project.org/).
Clinical profiles on clinical specimens and patients
Cancer tissues and their pair-matched adjacent normal tissues were obtained from patients diagnosed with CRC at the Affiliated Hospital of Nantong University. None of the patients had undergone chemotherapy or pre-operative radiotherapy. Written informed consent was obtained from all patients, and this study was approved by the Ethics Committee of the Affiliated Hospital of Nantong University (Approval number: 2019-L053).
Cell lines and cell culture
The human CRC cell lines (NCM460, CACO2, DLD1, SW1116, and SW620) were purchased from the Chinese Academy of Sciences (Shanghai, China). Authentication of each cell line was confirmed via short tandem repeat analysis, and the cells were cultured for fewer than 6 months after testing. All cell lines were maintained in RPMI 1640 medium (Thermo, NY, USA) supplemented with 10% fetal bovine serum (Gibco, CA, USA) and incubated at 37 °C in a 5% CO2 environment. Small interfering RNAs (siRNAs) targeting SLAMF9 were obtained from Ribobio (Guangzhou, China), and their sequences were as follows: SLAMF9-siRNA-1, 5′-GGCUUUACCAAGCUCAAGUTT-3′; SLAMF9-siRNA-2, 5′-CACUUAUACAUUCCAUGAATT-3′; SLAMF9-siRNA-3, 5′-CUAUGCAGAUCCUAACUAUTT-3′. Transfection was performed using lipoFiter 3.0 (Hanbio, Shanghai, China), following the manufacturer’s instructions. For mouse tumorigenesis studies, the siRNA was modified with 2′-O-Methyl (2′-OMe) and obtained from GenePharma (Shanghai, China).
Cell counting kit-8 (CCK-8) assay
Cell proliferation was assessed using the CCK-8 assay (Dojindo, Kumamoto, Japan). For this experiment, 3 × 103 cells/well in 100 μL of media were seeded in triplicate wells of 96-well plates at 48 h post-transfection. After 24, 48, 72 and 96 h, 10 μL of CCK-8 solution was added to each well and incubated for 2 h at 37 °C. Absorbance at 450 nm was measured using a microplate reader (Thermo, Waltham, MA, USA).
Migration and invasion assays
For migration assays, Transwell inserts (8-μm pore size) from Millipore (NY, USA) were utilized. A 24-well plate containing the upper chamber was filled with 100 μl/well of resuspended transfected cells in FBS-free RPMI 1640 media at a concentration of 5 × 105 cells/ml. The lower chambers were filled with 600 μl/well of RPMI 1640 with 10% FBS. The cells were then cultured for 48 h. Subsequently, the lower surface of the membrane’s infected cells was fixed, dyed, and photographed. Cell counts were performed by examining three randomly chosen microscope fields for each group. For invasion assays, Transwell membranes were coated with 50 μl of Matrigel (30 mg/well; BD Biosciences, USA) prior to conducting experiments in the same manner.
Immunohistochemistry (IHC)
Human CRC specimens underwent dewaxing and dehydration. Antigen retrieval was achieved by heating the sections in a microwave with sodium citrate buffer (10 mM, pH 6.0). The sections were incubated in a citric acid buffer for 20 min to complete the antigen retrieval process. Following this, the slides were treated with 0.3% H2O2, blocked with 5% goat serum, and then incubated overnight with a rabbit polyclonal antibody against SLAMF9 (1:400, Novus, USA). Visualization of the results was performed using 3,3′-diaminobenzidine tetrahydrochloride (DAB) staining. Following staining, the slides underwent a five-minute hematoxylin redye process. The immune complexes were observed under a microscope after undergoing cleaning, dehydration, transparency, and fixation using a gel. Each experiment was conducted in triplicate.
Western blotting analysis
Proteins were separated by 12% SDS-PAGE and transferred to PVDF membranes. The blots were then blocked in 5% evaporated milk at room temperature for 1.5 h and subsequently incubated with either rabbit polyclonal anti-human SLAMF9 antibody (1:1000, Novus, USA) or mouse anti-human β-tubulin antibody (1:1000, Abmart, China). After incubation, the membranes were washed and treated with the appropriate secondary antibody. Protein bands were visualized using an enhanced chemiluminescence system (ECL; Beyotime Institute of Biotechnology), and densitometry was used to quantify the protein bands after a second round of washing and incubation.
Tumor formation in nude mice
The animal experiments conducted in this study were approved by the Institutional Animal Care and Use Committee of the Affiliated Hospital of Nantong University (Approval number: S20211217-005). DLD1 cells were stably transfected, and nude mice (BALB/c Nude, 4 weeks old) were obtained from the Experimental Animal Centre of Nantong University. Each mouse was subcutaneously injected with 1 × 107 cells on the ventral side for tumor formation assays. Tumor volume on the ventral side was measured at 4, 8, 12, 16, 20, 24 and 28 days post-injection. Mice were sacrificed by cervical dislocation 4–5 weeks after injection, and tumor weight was measured. All experimental procedures were conducted following relevant guidelines and regulations and are reported in accordance with the ARRIVE guidelines.
Statistical analysis
Gene expression analysis was conducted using the student’s t-test. Survival analysis was performed utilizing Kaplan–Meier curves, the log-rank test, and the Cox proportional hazards regression model. Additionally, the Spearman test was employed to analyze TMB and MSI. These analyses were executed using the R statistical software package (version 4.0.3) and supplemented with R language software packages, including ggplot2, clusterProfiler, GSVA, and enrichPlot. A significance level of P < 0.05 was considered statistically significant.
Ethical approval
The present study was approved by the Ethics Committee of the Affiliated Hospital of Nantong University (Approval number: 2019-L053). All experiments adhered to the principles outlined in the Helsinki Declaration and institutional guidelines. Written informed consent was obtained from all patients for the use of their tissue for research purposes. Animal experiments were approved by the Experimental Animal Ethics Committee of Nantong University (Approval number: S20211217-005).
Results
SLAMF9 expression profiles in human normal and cancers tissues
To evaluate the abnormal expression of SLAMF9 in cancers, we analyzed its expression levels using the TCGA and GTEx data. In the TCGA data, SLAMF9 was found to be significantly upregulated in 16 of 20 cancer types, except in cholangiocarcinoma (CHOL), esophageal carcinoma (ESCA), pancreatic adenocarcinoma (PAAD), and prostate adenocarcinoma (PRAD) (Fig. 1A). However, when combining the data of TCGA and GTEx, significant differences were observed in 26 of 27 cancers (Fig. 1B). Next, we utilized the TCGA database to assess SLAMF9 mRNA expression levels in CRC patients and compared them with those in normal tissues. Our analysis revealed increased SLAMF9 mRNA expression in CRC tissues compared to normal tissues (Fig. 1C). These findings were further validated in CRC and paired adjancent normal tissues (Fig. 1D). Additionally, the area under the curve (AUC) for SLAMF9 expression in CRC was 0.913 (95% CI 0.83–0.944) (Fig. 1E). Overall, these results highlight the potential diagnostic implications of SLAMF9 mRNA expression in CRC patients.
SLAMF9 mRNA expression in CRC tissues. (A) Expression of SLAMF9 mRNA in pan-cancer on the TCGA database. (B) Combining the data from TCGA and GTEx, SLAMF9 expression differences between tumor and normal tissues. Expression level of SLAMF9 in normal tissues and paired adjacent tissues unmatched tissues (C) and matched tissues (D) in TCGA database. (E) The ROC curve analysis for SLAMF9 expression in CRC patients. ns, P ≥ 0.05; *P < 0.05; **P < 0.01; ***P < 0.001.
Examination of the association between SLAMF9 expression level and prognosis
To investigate the correlation between prognosis and SLAMF9 expression level, we assessed the OS and PFI of patients with each tumor. Utilizing data from the TCGA database, we assessed the association between SLAMF9 expression levels and OS across different tumor types by conducting Cox regression analysis. The results showed that SLAMF9 exhibited significant risk proportions for kidney chromophobe (KICH) (HR 1.35, 95% CI 1.03–1.77, P = 0.02), adrenocortical carcinoma (ACC) (HR 1.25, 95% CI 1.14–1.41, P = 5.6e−5), kidney renal clear cell carcinoma (KIRC) (HR 1.08, 95% CI 1.03–1.14, P = 1.0e−3), Lower Grade Glioma in the brain (LGG) (HR 1.28, 95% CI 1.21–1.35, P = 2.2e−17), lung adenocarcinoma (LUAD) (HR 1.10, 95% CI 1.02–1.19, P = 0.02), PAAD (HR 1.20, 95% CI 1.07–1.33, P = 1.5e−3), Pan-kidney cohort (KIPAN) (HR 1.05, 95% CI 1.01–1.09, P = 0.02), and colon adenocarcinoma (COAD) (HR 1.12, 95% CI 1.01–1.25, P = 0.04), with KICH showing the highest risk effect (Fig. 2A). Subsequent survival analysis, using patient information dichotomized by ideal cut-off values for each tumor type, revealed varying survival among OS-related tumor types, indicating poorer prognosis for patients with high SLAMF9 expression (Fig. 2B). Moreover, the Cox proportional hazards model demonstrated a significant association between SLAMF9 expression level and PFI in patients with ACC (HR 1.17, 95% CI 1.07–1.29, P = 3.5e−4), PAAD (HR = 1.18, 95% CI 1.06–1.30, P = 2.8e−3), KIPAN (HR 1.05, 95% CI 1.01–1.10, P = 0.01), LGG (HR 1.21, 95% CI 1.15–1.27, P = 4.4e−15), pheochromocytoma and paraganglioma (PCPG) (HR 1.15, 95% CI 1.02–1.30, P = 0.02), thyroid carcinoma (THCA) (HR 1.10, 95% CI 1.01–1.20, P = 0.03), Testicular Germ Cell Tumors (TGCT) (HR 1.14, 95% CI 1.01–1.28, P = 0.04), and KICH (HR 1.24, 95% CI 1.00–1.52, P = 0.04) (Fig. 2C). Furthermore, Kaplan–Meier survival analysis indicated that patients with high levels of SLAMF9 had shorter PFI in ACC (HR 3.15, 95% CI 1.67–5.94, P = 1.8e−4), GBMLGG (HR 3.51, 95% CI 2.78–4.45, P = 1.5e−28), KIPAN (HR 1.50, 95% CI 1.12–2.00, P = 6.0e−3), KIRC (HR 1.80, 95% CI 1.29–2.50, P = 4.4e−4), PAAD (HR 1.65, 95% CI 1.11–2.45, P = 0.01), and THCA (HR 2.08, 95% CI 1.04–4.17, P = 0.03) (Fig. 2D).
Survival analysis of SLAMF9 in pan-cancer. (A)The relationship between SLAMF9 expression levels and OS in various cancer types through single variate Cox regression analysis using data from the TCGA database. (B) The survival analyses for median expression value in various cancer types using data from the TCGA database. (C) Correlation between SLAMF9 expression and PFS analyzed by Cox regression using data from the TCGA database. (D) The survival analysis for SLAMF9 expression and prognosis in various cancer types.
Association between the expression of SLAMF9 and immune infiltrating level of tumors
To investigate the relationship between SLAMF9 expression and the tumor immune response, we utilized the TIMER database to assess immune infiltration in human tumors across different SLAMF9 expression levels. Overall, SLAMF9 exhibited a positive correlation with immune infiltrating levels of various components, including cancer-associated fibroblasts (CAFs), macrophages, myeloid-derived suppressor cells (MDSCs), monocytes, neutrophils, and natural killer (NK) cells. However, a negative correlation was observed between SLAMF9 expression and CD8+ T cells. This profile suggests that SLAMF9 plays a role in immune infiltration to some extent and is essential for immuno-oncological interactions (Fig. 3A).
To determine whether SLAMF9 contributes to the modulation of immune infiltration across multiple cancers, we assessed the association between SLAMF9 expression and cancer purity. As shown in Fig. 3B, SLAMF9 expression exhibited strong associations with stromal score, immune score, and ESTIMATE score, with the most significant correlations observed in COAD, rectum adenocarcinoma (READ), and GBMLGG.
Correlation between immunotherapy, immune checkpoints, and SLAMF9
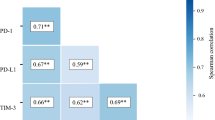
The pan-cancer associations between SLAMF9 and immunological checkpoints are shown in Fig. 4A. In several tumors such as CHOL, COAD, KICH, brain LGG, PCPG, READ, and thymoma (THYM), significant relationships were observed between SLAMF9 expression and the expression levels of well-known immune checkpoints such as neuronilin-1 (NRP1), leukocyte-associated immunoglobulin-like receptor-1 (LAIR1), CD48, CD28, HAVCR2, CD276, CD80, CD70, CD274, CD86, and CD44, suggesting potential cooperation between SLAMF9 and established immunological checkpoints.
Correlations between SLAMF9 and immune checkpoints, as well as other variables of interest. (A) The correlations between SLAMF9 and confirmed immune checkpoints in multiple cancers. (B) The correlations between SLAMF9 and essential genes involved in MMR in multiple cancers. (C,D) The correlations of SLAMF9 expression and TMB, MSI in cancers. (*P < 0.05, **P < 0.01, ***P < 0.001).
During DNA replication and genetic recombination, the mismatch repair mechanism plays a crucial role in detecting and correcting mismatched nucleotides18. MSI, characterized by frequent polymorphisms in short repetitive DNA sequences and single nucleotide substitutions, is a hypermutator phenotype resulting from DNA mismatch repair deficiency. This condition leads to an increased TMB by accumulating mutations in cancer-related genes19,20,21,22. MSI and TMB are considered independent predictors of immune checkpoint blockade (ICB) effectiveness and are involved in tumor initiation23,24,25. In this study, we investigated the relationship between SLAMF9 expression and several critical mismatch repair (MMR) characteristics. Our analysis revealed that SLAMF9 expression was negatively correlated with MutL homolog 1 (MLH1) in CHOL, HNSC, LGG, OV, and THYM. Conversely, in ACC, SLAMF9 expression was positively correlated with MutS homolog 2 (MSH2) and MutS homolog 6 (MSH6). However, there was a negative association between THYM and UCEC. In COAD, ESCA, GBM, head and neck squamous cell carcinoma (HNSC), KIRP, LGG, and THYM, it was negatively associated with epithelial cell adhesion molecule (EPCAM) (Fig. 4B). Additionally, in BRCA, cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), COAD, HNSC, LGG, Ovarian serous cystadenocarcinoma (OV), PAAD, sarcoma (SARC), skin cutaneous melanoma (SKCM), STAD, uterine corpus endometrial carcinoma (UCEC) and BLCA, we found that SLAMF9 expression demonstrated a favorable association with TMB (Fig. 4C). Most MSI-High tumors were found to express more SLAMF9 than genetically stable ones, while the opposite trend was observed for LUAD and lung squamous cell carcinoma (LUSC) cohorts (Fig. 4D).
Functional analysis by GSEA
To investigate the pathways associated with SLAMF9 in tumor immunosuppression, we determined the differentially expressed genes (DEGs) between SLAMF9 high-expression and low-expression groups. The results indicated a positive correlation between SLAMF9 expression and various immune-related pathways across different tumors, including TNFA-signaling-via-NFKB, inflammatory response, IL6-JAK-STAT3 signaling, IL2-STAT5 signaling, and epithelial–mesenchymal transition (EMT) (Fig. 5A). Notably, EMT plays a significant role in tumor metastasis, a leading cause of mortality26,27. To determine the association between SLAMF9 expression and tumor metastasis, we examined the correlation between SLAMF9 expression and EMT score. The analysis revealed a significant positive correlation between SLAMF9 expression and EMT score in most malignancies (Fig. 5B). Furthermore, GSEA was employed to assess the functional enrichment of DEGs. Generally, the top three positively enriched KEGG terms in the high SLAMF9 subgroup were pathogenic Escherichia coli infection, glycolysis gluconeogenesis, and pentose phosphate pathway. Conversely, negatively enriched terms included basal cell carcinoma. Additionally, the top positively enriched HALLMARK terms encompassed hypoxia, glycolysis, and EMT (Fig. 5C). These results suggest that the immunosuppressive microenvironment facilitated by SLAMF9 across various cancers could be guided by these pathways.
Clinical correlation analysis of SLAMF9 in colorectal carcinoma
Since the above findings suggest that SLAMF9 plays a key role in the development of CRC, we further analyzed the association between SLAMF9 expression in the TCGA database and clinical characteristics in CRC (Table 1). The results indicated that SLAMF9 expression level was significantly associated with T stage (P = 0.005), N stage (P = 0.007), and pathological stage (P = 0.016). Next, logistic analysis was employed to analyze the relationship between different clinical parameters and the prognosis of patients, revealing associations of the T stage, N stage, and pathological stage with OS (Table 2), and the results indicate that SLAMF9 expression was linked to the malignant pathological progression of CRC. For further confirmation of the role of SLAMF9 in CRC occurrence and development, the expression of SLAMF9 in CRC and surrounding tissues was initially examined. The results from western blotting indicated that SLAMF9 expression was increased in CRC tissues compared to para-cancerous tissues (Fig. 6A). Immunohistochemistry was utilized to assess SLAMF9 protein expression in CRC tissues, revealing an upregulation of SLAMF9 protein expression in CRC tissues (Fig. 6B). Subsequently, SLAMF9 mRNA expression in five CRC cell lines was examined using qRT-PCR and western blotting to further verify SLAMF9 expression in CRC. The results showed that compared with NCM460, the level of SLAMF9 was significantly increased in four CRC cells (Fig. 6C), which was consistent with the observed protein levels in western blotting (Fig. 6D).
Validation of SLAMF9 expression in CRC. (A) Comparing the SLAMF9 expression in paired normal and tumor by WB. (B) Representative immunohistochemical analysis of SLAMF9 in CRC, bar = 50 μm. (C) Relative mRNA expression of SLAMF9 in CRC cell lines. (D) Expression levels of SLAMF9 protein in CRC cell lines (*P < 0.05, **P < 0.01, ***P < 0.001).
SLAMF9 promotes cell viability and motility in CRC cells
To investigate the role of SLAMF9 in colorectal carcinogenesis, we used three siRNAs to knock down the expression of SLAMF9 in CRC cells. The efficacy of the knockdown transfection was assessed by qRT-PCR and western blotting, revealing a significant reduction in SLAMF9 expression in CRC cells following siRNA transfection (Fig. 7A). CCK‐8 assays demonstrated a significant decrease in cell proliferation ability in siRNA-transfected CRC cells compared to the siNC group (Fig. 7B). We conducted transwell migration experiments to explore the impact of SLAMF9 on CRC cell invasion and migration, finding that SLAMF9 was positively associated with the migration and invasive abilities of CRC cells (Fig. 7C). To ascertain whether SLAMF9 increases the invasiveness of CRC through mechanisms related to EMT, we identified EMT indicators via western blotting. We observed that E-cadherin, an epithelial cell marker, was significantly expressed in siSLAMF9-transfected CRC cells, while the expression of mesenchymal markers Vimentin and N-cadherin decreased in CRC cell lines transfected with siSLAMF9, indicating a shift towards a mesenchymal phenotype. Overall, our findings suggest that SLAMF9 induces EMT to facilitate tumor invasion and metastasis (Fig. 7D).
Effects of SLAMF9 knockdown on cell proliferation and migration. (A) RT-PCR and WB determine the efficiency of SLAMF9 knockdown in CRC cells. (B) The cells growth rates were determined by CCK-8 proliferation assays at various time points. (C) Transwell analysis was preformed to determine the cell migration. (D) The protein expression of E-cadherin, N-cadherin and vimentin in the indicated cells was examined by WB (*P < 0.05, **P < 0.01, ***P < 0.001).
SLAMF9 knockdown inhibits tumor growth in vivo
According to the findings indicating that SLAMF9 inhibits CRC cell proliferation, migration, and invasion in vitro, we proceeded to investigate its effect on CRC growth in vivo. Initially, we established a nude mice model by subcutaneously injecting DLD1 cells into the ventral side and monitored tumor growth regularly (Fig. 8A and B). Two weeks after subcutaneous injection, we observed significantly lower tumor weight and size in the SLAMF9 knockdown group compared to the control group (Fig. 8C and D). Immunohistochemical staining of tumor tissues further confirmed these results, showing lower expression of Ki-67 in the SLAMF9-siRNA group (Fig. 8E). Collectively, these in vivo findings suggest that SLAMF9 may function as an oncogene, promoting CRC cell proliferation, migration, and invasion.
Knockdown of SLAMF9 inhibits colorectal cancer cell tumorigenesis in vivo. (A) The appearance of nude mice from SLAMF9 NC and SLAMF9-si groups. (B) Tumor size of the two groups. (C) Tumor images of nude mice in the two groups. (D) Weight of tumors in the two groups. (E) The tumor sections were under IHC staining using antibodies against ki-67, bar = 50 μm (*P < 0.05).
Discussion
SLAMF comprises nine members, all located at the same locus28,29. Recently, SLAMF9 was discovered as a new member of the SLAMF family. While its intracellular tail lacks known signaling motifs, it does possess a transmembrane region, an extracellular amino-terminal variable immunoglobulin-like domain, and a membrane-proximal constant immunoglobulin-like domain, similar to other Slamf receptors. Studies have shown the presence of slamf9 mRNA in various immune cells, including human T cells, B cells, monocytes, and DCs30,31,32. Enrichment analysis has linked high SLAMF9 expression with immune response33. However, both a signal transduction mechanism and a ligand for SLAMF9 remain elusive. Unique effects of SLAMF8/9 deficiency in macrophages have been observed, but the precise role of SLAMF9 remains largely unknown. Given these considerations, we hypothesized that SLAMF9 may play a significant role in immune system regulation, motivating our focus on this gene.
Our findings indicate that SLAMF9 expression is significantly elevated in many cancers compared with normal tissues. High SLAMF9 levels could be likely necessary for the control of the immune response, which could account for the protein’s elevated expression in most tumor tissues. This increased expression was found to be associated with worse OS and PFI in those cancers. Under typical circumstances, the immune system is able to identify and eliminate tumor cells from the TME34,35,36. However, tumor cells may employ a number of strategies for surviving and proliferating while evading the immune system. Tumor immunotherapy, including immune checkpoint inhibitors of the monoclonal antibody family37, cancer vaccines38, therapeutic antibodies39, and cell therapy40, may restore the body’s innate ability to combat tumors. Clinical outcomes for various cancers are influenced by tumor-infiltrating immune cells (TIICs). Through the analysis of over 40 frequently occurring immune checkpoint genes, we identified a correlation between SLAMF9 expression and tumor purity, as well as an inverse relationship with TIICs, which suggests an association between SLAMF9 expression and tumor infiltration levels. Moreover, specific clinicopathological characteristics, such as increased TMB and lymphocytic infiltration, have been associated with MSI, which in turn is linked to elevated cancer risks. Notably, TMB has emerged as a latent biomarker for predicting ICB response41 with potential implications for immune-related survival outcomes in breast cancer patients42. Consequently, future investigations may use SLAMF9 expression levels to assess the impact of immunotherapy. Furthermore, the development of SLAMF9-targeted therapies could complement conventional immunotherapies, enhancing their effectiveness.
We also investigated the protein interactions and intracellular signaling control, both of which were found to be significant. Previous studies have shown that simultaneous knockdown of SLAMF8 and SLAMF9 can prevent endotoxin-induced liver inflammation by suppressing TLR4 expression in macrophages. Another study revealed that SLAMF9 regulates plasmacytoid DC (pDC) homeostasis and function in both healthy and pathological conditions, which aligns with our hypothesis, suggesting that SLAMF9 plays a key role in immune regulation. Furthermore, we examined the function of SLAMF9 in CRC using molecular biological techniques and observed upregulated SLAMF9 expression in CRC tissues, as confirmed by Western blotting and immunohistochemistry. CCK-8 assays demonstrated that SLAMF9 promoted CRC cell proliferation. Transwell experiments indicated that SLAMF9 enhanced CRC cell invasion and migration, and SLAMF9 was found to induce EMT, an essential process for cancer invasion and metastasis43. During EMT, cells acquire mesenchymal characteristics at the expense of epithelial traits. Notably, our study demonstrated that decreasing SLAMF9 expression suppressed EMT by increasing the expression of the epithelial marker E-cadherin and reducing the expression of mesenchymal markers such as vimentin and N-cadherin. Taken together, these findings support the validity and reliability of the bioinformatics study results in CRC. In the future, we aim to conduct similar molecular biological validations in additional malignancies.
Despite our comprehensive exploration and integration of data from various databases, our present study still had some limitations that should be clarified. While molecular biology techniques confirmed that SLAMF9 promotes cancer in CRC and the bioinformatic analysis provided valuable insights into SLAMF9’s role in malignancies, further in vitro or in vivo biological experiments are necessary to validate our findings and improve their therapeutic applicability. Additionally, it remains uncertain whether SLAMF9 affects clinical survival through immunological mechanisms despite its association with immunity and clinical outcomes in human malignancies.
Conclusion
In summary, our research highlights the key role of SLAMF9 in tumor development and metastasis. Based on the obtained results, we proposed a potential mechanism by which SLAMF9 influences tumor immunology, metabolic activity, and EMT in various cancers. Future studies focusing on SLAMF9 expression and its interaction with the tumor immune microenvironment could pave the way for the development of immunotherapy-based anti-cancer strategies.
Data availability
The original available statements demonstrated in the study are summarized in the article materials. Additional inquiries should be submitted to the corresponding authors.
References
Tang, J., Pearce, L., O’Donnell-Tormey, J. & Hubbard-Lucey, V. M. Trends in the global immuno-oncology landscape. Nat. Rev. Drug Discov. 17(12), 922 (2018).
Kim, J. et al. Hierarchical contribution of individual lifestyle factors and their interactions on adenomatous and serrated polyp risk. J. Gastroenterol. 58(9), 856–867 (2023).
Park, K. H. et al. Genomic landscape and clinical utility in Korean advanced pan-cancer patients from prospective clinical sequencing: K-MASTER program. Cancer Discov. 12(4), 938–948 (2022).
Ju, M. et al. Pan-cancer analysis of NLRP3 inflammasome with potential implications in prognosis and immunotherapy in human cancer. Brief. Bioinform. 22(4), bbaa345 (2021).
Bagaev, A. et al. Conserved pan-cancer microenvironment subtypes predict response to immunotherapy. Cancer Cell 39(6), 845-865 e7 (2021).
Dragovich, M. A. & Mor, A. The SLAM family receptors: Potential therapeutic targets for inflammatory and autoimmune diseases. Autoimmun. Rev. 17(7), 674–682 (2018).
Zeng, X. et al. Combined deficiency of SLAMF8 and SLAMF9 prevents endotoxin-induced liver inflammation by downregulating TLR4 expression on macrophages. Cell Mol. Immunol. 17(2), 153–162 (2020).
von Wenserski, L. et al. SLAMF receptors negatively regulate B cell receptor signaling in chronic lymphocytic leukemia via recruitment of prohibitin-2. Leukemia 35(4), 1073–1086 (2021).
Mak, A. et al. Brief report: Decreased expression of CD244 (SLAMF4) on monocytes and platelets in patients with systemic lupus erythematosus. Clin. Rheumatol. 37(3), 811–816 (2018).
Teng, L. et al. SLAMF8 participates in acute renal transplant rejection via TLR4 pathway on pro-inflammatory macrophages. Front. Immunol. 13, 846695 (2022).
Goldman, M. J. et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 38(6), 675–678 (2020).
Dong, Q. et al. MARCO is a potential prognostic and immunotherapy biomarker. Int. Immunopharmacol. 116, 109783 (2023).
Li, T. et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 48(W1), W509–W514 (2020).
Shen, S. et al. Development and validation of an immune gene-set based prognostic signature in ovarian cancer. EBioMedicine 40, 318–326 (2019).
Tang, Z. et al. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45(W1), W98–W102 (2017).
Powers, R. K., Goodspeed, A., Pielke-Lombardo, H., Tan, A. C. & Costello, J. C. GSEA-InContext: Identifying novel and common patterns in expression experiments. Bioinformatics 34(13), i555–i564 (2018).
Xu, W. X. et al. An integrative pan-cancer analysis revealing LCN2 as an oncogenic immune protein in tumor microenvironment. Front. Oncol. 10, 605097 (2020).
Petrelli, F., Ghidini, M., Ghidini, A. & Tomasello, G. Outcomes following immune checkpoint inhibitor treatment of patients with microsatellite instability-high cancers: A systematic review and meta-analysis. JAMA Oncol. 6(7), 1068–1071 (2020).
Harmsen, T., Klaasen, S., van de Vrugt, H. & Te Riele, H. DNA mismatch repair and oligonucleotide end-protection promote base-pair substitution distal from a CRISPR/Cas9-induced DNA break. Nucleic Acids Res. 46(6), 2945–2955 (2018).
Bielska, A. A. et al. Tumor mutational burden and mismatch repair deficiency discordance as a mechanism of immunotherapy resistance. J. Natl. Compr. Canc. Netw. 19(2), 130–133 (2021).
Huang, Q. R. et al. The prognostic and immunological role of MCM3 in pan-cancer and validation of prognosis in a clinical lower-grade glioma cohort. Front. Pharmacol. 15, 1390615 (2024).
Xu, Y. et al. Microsatellite instability in mismatch repair proficient colorectal cancer: Clinical features and underlying molecular mechanisms. EBioMedicine 103, 105142 (2024).
Ho, W. W. et al. Dendritic cell paucity in mismatch repair-proficient colorectal cancer liver metastases limits immune checkpoint blockade efficacy. Proc. Natl. Acad. Sci. U S A 118(45), e2105323118 (2021).
Wang, Z. et al. Plasma-based microsatellite instability detection strategy to guide immune checkpoint blockade treatment. J. Immunother. Cancer 8(2), e001297 (2020).
Chen, Y. et al. Photoactivated formation of an extravascular dynamic hydrogel as an intelligent blood flow regulator to reprogram the immunogenic landscape. Nano Lett. 24, 5690 (2024).
Youssef, K. K. & Nieto, M. A. Epithelial-mesenchymal transition in tissue repair and degeneration. Nat. Rev. Mol. Cell Biol. 29, 1 (2024).
Yang, J. et al. Single-cell profiling reveals molecular basis of malignant phenotypes and tumor microenvironments in small bowel adenocarcinomas. Cell Discov. 8(1), 92 (2022).
Zhou, T., Guan, Y., Sun, L. & Liu, W. A review: Mechanisms and molecular pathways of signaling lymphocytic activation molecule family 3 (SLAMF3) in immune modulation and therapeutic prospects. Int. Immunopharmacol. 133, 112088 (2024).
Kloc, D. et al. SLAM family receptors in B cell chronic lymphoproliferative disorders. Int. J. Mol. Sci. 25(7), 4014 (2024).
Mikulin, J. A., Bates, B. L. & Wilson, T. J. A simplified method for separating renal MPCs using SLAMF9. Cytometry A 99(12), 1209–1217 (2021).
Christensen, S. M. et al. Host and parasite responses in human diffuse cutaneous leishmaniasis caused by L. amazonensis. PLoS Negl. Trop. Dis. 13(3), e0007152 (2019).
Wilson, T. J. et al. Signalling lymphocyte activation molecule family member 9 is found on select subsets of antigen-presenting cells and promotes resistance to Salmonella infection. Immunology 159(4), 393–403 (2020).
Sever, L. et al. SLAMF9 regulates pDC homeostasis and function in health and disease. Proc. Natl. Acad. Sci. U S A 116(33), 16489–16496 (2019).
Mao, X. et al. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: New findings and future perspectives. Mol. Cancer 20(1), 131 (2021).
Mendez-Gomez, H. R. et al. RNA aggregates harness the danger response for potent cancer immunotherapy. Cell 187, 2521 (2024).
Kaymak, I., Williams, K. S., Cantor, J. R. & Jones, R. G. Immunometabolic interplay in the tumor microenvironment. Cancer Cell 39(1), 28–37 (2021).
Bagchi, S., Yuan, R. & Engleman, E. G. Immune checkpoint inhibitors for the treatment of cancer: Clinical impact and mechanisms of response and resistance. Annu. Rev. Pathol. 16, 223–249 (2021).
Sahin, U. & Tureci, O. Personalized vaccines for cancer immunotherapy. Science 359(6382), 1355–1360 (2018).
Blanco, B., Dominguez-Alonso, C. & Alvarez-Vallina, L. Bispecific immunomodulatory antibodies for cancer immunotherapy. Clin. Cancer Res. 27(20), 5457–5464 (2021).
He, J. et al. Defined tumor antigen-specific T cells potentiate personalized TCR-T cell therapy and prediction of immunotherapy response. Cell Res. 32(6), 530–542 (2022).
Thomas, A. et al. Tumor mutational burden is a determinant of immune-mediated survival in breast cancer. Oncoimmunology 7(10), e1490854 (2018).
Chan, T. A. et al. Development of tumor mutation burden as an immunotherapy biomarker: Utility for the oncology clinic. Ann. Oncol. 30(1), 44–56 (2019).
Pastushenko, I. & Blanpain, C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 29(3), 212–226 (2019).
Acknowledgements
We sincerely appreciate all lab members.
Funding
This work is supported by the National Natural Science Foundation of China (No. 81972015) and by Fund Program of Social Development Project from Jiangsu Provincial Department of Science and Technology (No. BE2020770), the Nantong Science and Technology Program (MS12021003).
Author information
Authors and Affiliations
Contributions
Chunmei Zhao, Xingjia Zhu, and Huimin Liu signed and wrote the manuscript. Qingyu Dong, Jing Sun, Baolan Sun performed the data analysis. Guihua Wang and Xudong Wang helped the revision and validation. All authors contributed to this article and approved the submitted version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhao, C., Zhu, X., Liu, H. et al. The prognostic and immune significance of SLAMF9 in pan-cancer and validation of its role in colorectal cancer. Sci Rep 14, 17899 (2024). https://doi.org/10.1038/s41598-024-68134-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-68134-y
- Springer Nature Limited