Abstract
Although circular RNAs (circRNA) have been demonstrated to modulate tumor initiation and progression, their roles in the proliferation of hepatocellular carcinoma (HCC) are still poorly understood. Based on the analysis of GEO data (GSE12174), hsa-circRNA-0015004 (circ-0015004) was screened and validated in 80 sets of HCC specimens. Subcellular fractionation analysis was designed to determine the cellular location of circ-0015004. Colony formation and cell counting kit-8 were performed to investigate the role of circ-0015004 in HCC. Dual-luciferase reporter gene assays, RNA immunoprecipitation and chromatin immunoprecipitation were employed to verify the interaction among circ-0015004, miR-330-3p and regulator of chromatin condensation 2 (RCC2). The expression level of circ-0015004 was significantly upregulated in HCC cell lines and HCC tissues. HCC patients with higher circ-0015004 levels displayed shorter overall survival, and higher tumor size and TNM stage. Moreover, knockdown of circ-0015004 significantly reduced HCC cell proliferation in vitro and inhibited the growth of HCC in nude mice. Mechanistic studies revealed that circ-0015004 could upregulate the expression of RCC2 by sponging miR-330-3p, thereby promoting HCC cell proliferation. Furthermore, we identified that Ying Yang 1 (YY1) could function as an important regulator of circ-0015004 transcription. This study systematically demonstrated the novel regulatory signaling of circ-0015004/miR-330-3p/RCC2 axis in promoting HCC progression, providing insight into HCC diagnosis and treatment from bench to clinic.
Similar content being viewed by others
Introduction
Hepatocellular carcinoma (HCC), ranking as the third leading cause of cancer-related deaths in the world, accounts for about 90% of all cases of primary liver cancers1,2,3. Its primary etiological factors encompass cirrhosis and HBV infection4. Despite notable process in alternative therapies, surgical resection is still the most important way to treat HCC5, albeit with a dishearteningly low 5-year survival6. Hence, it is urgent to investigate the molecular mechanism of HCC and explore the novel potential biomarkers and therapeutic targets for HCC.
Circular RNAs (circRNAs), a subset of non-coding RNA molecules that primarily originate from pre-mRNA's reverse splicing process7, have been implicated in many stages of tumor progression, including tumor apoptosis, differentiation, metastasis, and proliferation8,9. In HCC, numerous studies have investigated the role of circRNAs in HCC proliferation. For instance, Xia et al. discovered that m6A-modified circMDK promotes HCC proliferation. Targeted nanotherapy against circMDK has been shown to inhibit HCC proliferation10. It's noteworthy that circ-ZEB1 was shown to play an important role in reinforcing the advancement of HCC, and the circ_0000098/miR-383/MCUR1 axis has been instrumental in orchestrating tumorigenesis in HCC11,12. Nevertheless, the precise roles and underlying mechanisms of circRNAs in HCC remain largely unknown. CircRNAs exert their biological functions through three main mechanisms: acting as miRNA sponges, interacting with RBPs, and translating proteins. The specific mechanism by which a circRNA functions can influence its ultimate biological role. Among them, circ-0015004, stemming from the succinate dehydrogenase C (SDHC) gene, situated on chr1:161293403–161326630, has been found to drive proliferation, invasion, and migration in renal cell carcinoma13. However, its role in HCC remains enigmatic.
Small RNA molecules known as microRNAs (miRNAs), characterized by their short length comprising 22 nucleotides, have the potential to exert either oncogenic or suppressive effects on tumors14. There is an emerging acknowledgment regarding circRNAs acting as competitive endogenous RNAs (ceRNAs), possessing the capability to influence the expression of subsequent miRNAs15. Within the context of HCC, the circRPN2 functions to restrain cell proliferation and glycolysis through modulation of the miR-183-5p/FOXO1 axis16. Similarly, the suppression of hsa_circRNA_104348 has been shown to hinder the advancement of HCC through targeting the miR-187-3p/RTKN2 axis17. Furthermore, miRNAs have exhibited their capacity to modulate mRNA levels by engaging with the 3′-untranslated regions (UTRs)18.
In this study, based on the analysis of GEO data (GSE12174), circRNA-0015004 was screened and validated in HCC specimens. We demonstrate that Ying Yang 1 (YY1)-regulated circRNA-0015004 promotes hepatocellular carcinoma development via miRNA-330-3p/regulator of chromatin condensation 2 (RCC2) axis.
Materials and methods
Human HCC tissue
This study was approved by the Medical Ethics Committee of Wujin Traditional Chinese Medicine Hospital, conformed to the Declaration of Helsinki. Informed consent was available for each patient. Tissue specimens were collected from 80 patients who underwent surgical procedures between March 2018 and June 2020. None of these individuals had undergone any preoperative chemotherapy or radiotherapy treatments. Tissue samples were meticulously collected and promptly preserved in liquid nitrogen for subsequent analysis. In parallel, an all-encompassing compilation of clinicopathological data and survival outcomes for all individuals involved was meticulously collated.
Cell culture
The HCC cell lines, including HCC-LM3, MHCC-97H, Hep3B, HepG2, and Huh-7, and the normal liver cell line L02, were purchased from the Shanghai Academy of Sciences. The cultivation of all cellular varieties was meticulously administered at a temperature of 37 °C, employing DMEM medium (Gibco, NY, USA) supplemented with 10% concentration of fetal bovine serum (Gibco), in addition to 100 mg/mL of streptomycin and 100 U/mL of penicillin. Subsequently, the cultured cells were maintained within a regulated environment infused with 5% CO2, fostering an optimum milieu for their expansion and proliferation.
RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR)
For total RNA extraction, a procedure utilizing Trizol reagent was executed for both tissue samples and cellular specimens. The extracted RNA underwent a subsequent reverse transcription process, yielding single-stranded cDNA. Following this, a thorough analysis via qRT-PCR was conducted using the SYBR Green Master Mix (Takara, Tokyo, Japan). GAPDH functioned as the intrinsic reference for both mRNA and circRNA analysis, whereas U6 was exclusively utilized as the internal reference for mRNA quantification. The 2−ΔΔCT method was used for precise computation of the expression level relative to the reference levels. Detailed information regarding the primers utilized can be found in Table S1.
Western blot analysis
Protein extraction from cells or tissues was performed using RIPA lysis buffer. The extracted proteins were subjected to separation via 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis, followed by transfer to a polyvinylidene fluoride membrane (Bio-Rad, CA, USA). To detect protein expression levels, a rabbit anti-RCC2 (CST; 3667S) polyclonal antibody was employed, with anti-GAPDH (CST; 92310SF) utilized as an internal control. The utilization of HRP-conjugated secondary antibodies (CST; 7074S) in conjunction with ECL Plus from EMD Millipore in Billerica, MA, USA, facilitated the precise detection of protein expression levels.
Experiments involving RNase R treatment
Following the amalgamation of RNase R and RNA acquired from Geneseed Biotech in Guangzhou, China, the composite underwent an incubation period of 20 min at 37 °C. Subsequently, an additional series of qRT-PCR analyses was undertaken to quantitatively assess the mRNA levels of both GAPDH expression and circ-0015004. This comprehensive procedure enabled a precise assessment of the targeted RNA molecules, offering valuable insights into their expression profiles.
Subcellular localization
To elucidate the subcellular localization of circ-0015004, we employed a PARIS Kit sourced from Invitrogen in Carlsbad, CA, USA. This kit facilitated a precise determination of the cellular compartment where circ-0015004 predominantly resides, offering crucial insights into its functional role.
Cell transfection
siRNAs for targeting circ-0015004 (siCirc1, siCirc2) and YY1 (si-YY1), and a negative control siRNA (NC) (RiboBio, Guangzhou, China) were transfected into MHCC and Hep3B cells. After 48-h transfection, the transfection efficacy was determined by qRT-PCR analysis. In addition, si-miR-330-3p, miR-330-3p mimics and their corresponding NC counterparts (RiboBio, Guangzhou, China) were transfected into the cells cells, by a transfection reagent (Lipofectamine 3000, Invitrogen). The specific sequences employed for these transfections are specified in Table S2.
CCK-8 assays
Following a 48-h transfection duration, the assessment of cellular proliferation was performed using a Cell Counting Kit-8 (CCK-8) (Vazyme, Nanjing, China). The cells were evenly distributed into a 96-well plate (Invitrogen), with a seeding density of 1 × 103 cells per well. Subsequent to seeding, measurements were taken at 450 nm optical density (OD) at intervals of 24, 48, 72, 96, and 120 h. To initiate the detection process, each well was treated with 10 μL of the CCK-8 reagent, followed by an additional 1-h incubation period at 37 °C.
Colony formation assays
Following resuspension, the transfected cells were evenly distributed onto six-well plate (Invitrogen), with each well containing 500 cells. Then, the cells were allowed to culture under continuous temperature conditions for a period spanning 10 days. Post-culture, the cells underwent three washes with PBS, followed by fixation using methanol for 10 min. Subsequent staining with 0.1% crystal violet was carried out, allowing for photographic documentation and subsequent cell counting.
Bioinformatics analysis
The GSE12174 dataset constitutes a gene expression library containing information on circRNA expression levels in normal liver tissue and liver cancer tissue. This dataset was obtained from the Gene Expression Omnibus (GEO) database, a publicly accessible repository dedicated to storing and sharing genetic data.
To analyze the dataset, the GEO2R online tool was used. GEO2R is a web-based tool that provides a simple and user-friendly interface for analyzing gene expression data.
In this instance, the screening criteria employed adjusted P value < 0.05 and included logFC < 1. These specific criteria were utilized to pinpoint dysregulated circRNAs, which denote circRNAs exhibiting either upregulation or downregulation in HCC tissues compared to normal liver tissues.
Subsequently, the Starbase database was utilized to forecast the target genes of two dysregulated circRNAs identified in the GSE12174 dataset: circ-0015004 and miR-330-3p. Starbase is a web-based platform that provides a variety of tools for predicting the functions and targets of circRNAs. In this case, it was used to identify the genes that are likely to be regulated by these circRNAs.
Finally, the JASPAR database was used to predict possible upstream regulators of the dysregulated circRNAs. JASPAR is a database of transcription factor binding profiles, which are used to predict the regulatory elements in the genome. In this case, it was used to identify transcription factors that may be involved in the regulation of the circRNAs and their target genes.
Dual-luciferase reporter gene assays
For predicting binding sites, we utilized online bioinformatics platforms (Circinteractome, miRwalk). Following this, elaborately engineered recombinant luciferase vectors encompassing both mutant (MUT) and wild-type (WT) sequences of circ-0015004 and RCC2 were constructed. These sequences were subsequently integrated into the pMIR-REPORT vector obtained from Ambion in Waltham, MA, USA. Co-transfection experiments were subsequently conducted in MHCC and Hep3B cells using the negative control (NC), miR-330-3p mimics, or si-YY1, assisted by Lipofectamine 3000 from Invitrogen. Following a 48-h incubation, the luciferase activity was assessed utilizing the dual-luciferase reporter gene detection system obtained from Promega Corporation.
Examination employing RNA immunoprecipitation (RIP) analysis
For the conduction of RIP analysis, a RIP kit was procured from Merck in Darmstadt, Germany. Briefly, MHCC and other cells were lysed using buffer for RIP lysis. The lysates were subsequently exposed to magnetic beads that had been previously attached to either the anti-AGO2 or anti-IgG antibody. This incubation was performed at 4 °C for 6 h. Then, the magnetic beads underwent a thorough washing process followed by digestion with proteinase K. Subsequent scrutiny of the purified RNA was carried out through qRT-PCR analysis, providing valuable insights into the interactions between the target molecules and AGO2.
Chromatin immunoprecipitation (ChIP)
The ChIP assay was conducted using a ChIP Chromatin Immunoprecipitation Kit acquired from Millipore in Massachusetts, USA. Formaldehyde was used to initiate cross-linking within the chromatin, leading to its fragmentation via sonication into smaller sections. Following this fragmentation process, magnetic beads, pre-coated with either anti-YY1 (CST; 46395S) or anti-IgG (CST; 3900S) antibodies, were employed for immunoprecipitation. Following the collection of enriched chromatin fragments and the removal of cross-linking, a subsequent qRT-PCR analysis was conducted.
Statistical analysis
Data in this study were statistically assessed and graphed by using GraphPad Prism 8 (La Jolla, CA, USA). All values were recorded as the mean ± standard deviation (s.d.). All experiments were repeated three or more independent biological replicates. Statistical significance between the means of two groups was determined using Student’s t tests (normal distribution). The statistics of the means of multiple groups were performed using one-way ANOVA or two-way ANOVA. For survival analysis, the overall survival (OS) rate was calculated employing the Kaplan–Meier method and compared using the log-rank test. The correlation between circ-0015004 and miR-330-3p expression levels was determined using Pearson’s test (r, P). For analyzing the relationship between circ-0015004 expression and clinicopathological parameters, the P value was determined using the Pearson chi-square test. Statistical significance was denoted by *P < 0.05, **P < 0.01, and ***P < 0.001. These levels were considered indicative of significant differences in the observed data.
Methods statement
The experiments have approved by the institutional and licensing committee, including any relevant details. The article has confirmed that all experiments were performed in accordance with relevant guidelines and regulations.
Ethics approval and consent to participate
This study received approval from the Ethics Committee of Wujin traditional Chinese medicine hospital.
Results
The circ-0015004 level is upregulated in HCC tissues and HCC cell lines
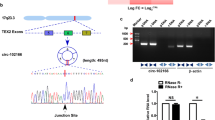
Dysregulated circRNAs in HCC tissues were identified through an assessment of the GSE12174 dataset. Based on the criteria of absolute log-fold change (|logFC|) and adjusted P value, we observed a substantial elevation in circ-0015004 expression in HCC tissues (Fig. 1A). In accordance with data acquired from the UCSC Genome Browser, circ-0015004 originated from the back-splicing process involving exons 2, 3, 4, and 5 within the SDHC gene (Fig. 1B). Then, an assessment employing qRT-PCR was conducted on a cohort consisting of 80 individuals diagnosed with HCC. We observed a remarkable upregulation in the expression levels of circ-0015004 in HCC tissues compared to controls (Fig. 1C). To ascertain the clinical significance of the elevated expression of circ-0015004, a comprehensive analysis of the clinicopathological features among HCC patients was carried out. By employing the median expression level as the dividing threshold, patients were categorized into two distinct segments: a high-expression cohort consisting of 40 samples and a low-expression cohort consisting of 40 samples. According to the data presented in Table 1, the levels of circ-0015004 expression exhibited variations in accordance with the tumor size (P = 0.0441) and TNM stage (P = 0.0132). Nevertheless, there was no notable correlation observed with respect to age, gender, HBV infection, or liver cirrhosis. Moreover, individuals manifesting escalated circ-0015004 expression levels displayed a less promising prognosis when compared to those exhibiting lower expression levels (Fig. 1D). Additionally, circ-0015004 was highly expressed in all five HCC cell lines (HepG2, MHCC-97H, Huh7, Hep3B, and HCC-LM3) (Fig. 1E). After undergoing RNase R treatment, the circ-0015004 expression levels in MHCC-97H and Hep3B cells remained relatively unaltered, suggesting the stable presence of circ-0015004 in these cells (Fig. 1F,G). These findings strongly imply the potential role of circ-0015004 in HCC.
The elevation of circ-0015004 in HCC cell lines and tissues. (A) GSE12174 dataset was analyzed and circ-0015004 was selected as the research object. (B) Circ-0015004 was formed through the back-splicing of the 2, 3, 4 and 5 exons of the SDHC gene. (C) Expression of circ-0015004 in HCC tissues. (D) Survival analysis of HCC patients. (E) Expression analysis of circ-0015004 in L02 cell lines (F,G) and HCC verified the levels of circ-0015004 expression in MHCC-97h and Hep3B cell lines post RNase R treatment. **P < 0.01, ***P < 0.001.
Downregulation of circ-0015004 inhibits HCC cell proliferation
Given the elevated expression of circ-0015004 in MHCC-97H and Hep3B cells, we focused on these lines for subsequent in vitro assessments. QRT-PCR analysis revealed cytoplasmic predominance of circ-0015004 (Fig. 2A). After transfection with si-NC, si-Circ1, and si-Circ2, both si-Circ2 and si-Circ1 significantly suppressed the expression of circ-0015004 (Fig. 2B). Remarkably, the reduction of circ-0015004 impeded cellular proliferation (Fig. 2C). Furthermore, circ-0015004 knockdown markedly restrained the colony-forming capability of MHCC-97H and Hep3B cells (Fig. 2D). These results demonstrate that downregulation of circ-0015004 inhibits HCC cell proliferation.
Downregulation of circ-0015004 inhibits HCC cell proliferation. (A) An investigation into the localization of circ-0015004 within Experiments targeting the nuclear and cytoplasmic localization in MHCC-97h and Hep3B cells. (B) The levels of circ-0015004 expression in MHCC-97h and Hep3B cells following transfection. (C,D) CCK-8 and colony formation assays were conducted to evaluate the influence of circ-0015004 regarding cell proliferation in MHCC-97h and Hep3B cells. **P < 0.01, ***P < 0.001.
Circ-0015004 acted as a sponge for miR-330-3p
The target miRNAs of circ-0015004 were predicted by the circinteractome database. As shown in Fig.3A, there was a potential binding site between miR-330-3p and circ-0015004. Furthermore, miR-330-3p upregulation inhibited the luciferase activity of the circ-0015004 reporter plasmids, while there were no significant changes in luciferase activities under miR-330-3p binding site mutation (Fig. 3B). Moreover, RIP assays exhibited a more significant amplification of circ-0015004 and miR-330-3p during AGO2 immunoprecipitation compared to IgG immunoprecipitation (Fig. 3C). Additionally, a reciprocal relationship was detected between the expression levels of miR-330-3p and circ-0015004 in both HCC cell lines and tissue. There was a significant negative correlation between miR-330-3p and circ-0015004 expression levels (Fig. 3D–F). Notably, circ-0015004 knockdown significantly enhanced miR-330-3p expression in MHCC-97H and Hep3B cells (Fig. 3G). Taken together, these results demonstrate that circ-0015004 functioned as a sponge for miR-330-3p.
Circ-0015004 acted as a sponge for miR-330-3p. (A) Computational tools were employed to identify the sites where circ-0015004 and miR-330-3p complementarily bind. (B) A luciferase reporter examination uncovered miR-330-3p as a target of circ-0015004 in Hep3B and MHCC-97 h cells. (C) The assessment of circ-0015004 and miR-330-3p expression levels in MHCC-97 h and Hep3B cells was conducted via RIP analysis. (D) Relative miR-330-3p expression in HCC tissues compared to adjacent normal tissues. (E) Relative miR-330-3p expression in L02 and HCC cell lines. (F) The correlation between circRNA-0015004 and miR-330-3p. (G) The mRNA expression level of miR-330-3p. **P < 0.01, ***P < 0.001.
MiR-330-3p impedes HCC cell proliferation
To uncover the governing process by which miR-330-3p influences the proliferation of HCC cells, we transfected miR-330-3p mimics into MHCC-97H and Hep3B cells. The efficacy of transfection was confirmed by qRT-PCR analysis (Fig. 4A). CCK-8 assays and colony formation analyses unequivocally showed that the enforced expression of miR-330-3p significantly restrained the growth capacity of HCC cells (Fig. 4B,C).
MiR-330-3p impedes HCC cell proliferation. (A) Expression levels of miR-330-3p were comparatively analyzed in transfected MHCC-97H and Hep3B cells. (B,C) To assess the impact of miR-330-3p on cell proliferation in both MHCC-97h and Hep3B cells, CCK-8 and colony formation assays were utilized. *P < 0.05, **P < 0.01, ***P < 0.001.
RCC2 acts a direct downstream target of miR-330-3p
To explore the regulatory mechanism of miR-330-3p in HCC cells, we employed miRwalk, miRtarbase, and miRDB to identify potential candidates for the target genes of miR-330-3p (Fig. 5A). Among the six candidate targets, RCC2 emerged prominently. RCC2 mRNA levels exhibited significant unregulation in both HCC cell lines and tissues (Fig. 5B,C). Analysis from the Kaplan–Meier Plotter database unveiled an association between elevated RCC2 expression and a poorer prognosis (Fig. 5D). Moreover, MHCC-97H and Hep3B cells were transfected with the pMIR-REPORT luciferase vector containing both WT) and mutant RCC2-3′ UTR sequences, alongside either miR-330-3p mimics or a negative control. As shown in Fig. 5E–G, there was a notable reduction in fluorescence intensity in the WT group after the transfection with miR-330-3p mimics, whereas there was no significant alteration in the mutant group. Circ-0015004 knockdown decreased the RCC2 expression (Fig. 5H,J). Additionally, elevated expression of miR-330-3p led to a noteworthy decrease in both RCC2 mRNA and protein levels (Fig. 5I,K). Conversely, the reduction of RCC2 triggered by the inhibition of circ-0015004 was counteracted upon miR-330-3p knockdown (Fig. 5L–O). Collectively, these results suggest that circ-0015004 upregulates RCC2 expression by decrease the expression of miR-330-3p.
RCC2 acts a direct downstream target of miR-330-3p. (A) Computational tools such as MiRDB, miRtarbase, and miRwalk were harnessed to pinpoint potential gene targets susceptible to the impact of miR-330-3p. (B,C) A comparative analysis conducted for the purpose of evaluating RCC2’s distinct expression in both HCC cell lines and tissues. (D) Analysis from the Kaplan–Meier Plotter database illuminates the association of elevated RCC2 expression with a more adverse prognosis. (E) Employing computational tools, potential sites for binding between miR-330-3p and RCC2 were identified. (F,G) Employed a luciferase reporter gene test highlighted the interaction between RCC2 in MHCC-97h and Hep3B cells and miR-330-3p. (H,I) The relative mRNA expression levels of RCC2 were assessed in Hep3B and MHCC-97h cells post transfection. (J,K) The assessment of RCC2 protein expression levels was carried out in transfected MHCC-97h and Hep3B cells. (L,M) The relative mRNA expression levels of RCC2 were assessed in Hep3B and MHCC-97h cells post transfection. (N,O) The assessment of RCC2 protein expression levels was carried out in transfected MHCC-97h and Hep3B cells. **P < 0.01, ***P < 0.001.
YY1 functions as a transcriptional regulator of circ-0015004
To investigate the transcriptional regulation mechanism of circ-0015004 in HCC, we used JASPAR database to explore the potential transcription factors of circ-0015004 (Fig. 6A). Among these candidates, our focus zeroed in on YY1 for further investigation. The ChIP assay was applied to examine the enrichment of YY1 onto the circ-0015004 promoter, indicating the binding of YY1 on circ-0015004 promoter (Fig. 6B). Additionally, the dual-luciferase reporter gene assay showed that si-YY1 markedly diminished the fluorescence intensity in the WT group (Fig. 6C). Furthermore, YY1 knockdown significantly decreased the expression of circ-0015004 (Fig. 6D,E). Collectively, these results demonstrate the role of YY1 serving as a crucial regulator for circ-0015004 transcription.
YY1 acts as a transcriptional regulator of circ-0015004. (A) Examination of the Jaspar database unveiled the prospective binding locus of YY1 within the promoter region of circ-0015004. (B) Results from the ChIP assay revealed the occupancy of YY1 in the promoter segment of circ-0015004. (C) Assessment of comparative luciferase activity in both Hep3B and MHCC-97 h cells. (D,E) Examination of the comparative levels of expression regarding rcc2 and circ-0015004 in Hep3B and MHCC-97h cells. **P < 0.01, ***P < 0.001.
Suppressing the expression of circ-0015004 inhibits HCC growth in vivo
Given downregulation of circ-0015004 inhibits HCC cell proliferation, we further examined the effect of circ-0015004 on tumour growth in vivo. The results showed that circ-0015004 knockdown markedly decreased tumour volume and weight compared to the controls (Fig. 7). These results suggested that circ-0015004 was involved in HCC growth in vivo.
Discussion
In recent years, circRNA has been increasingly researched in various fields, such as tumors19, autoimmune diseases20, and inflammatory diseases21. Its role in tumors has garnered significant attention, particularly in HCC22, colon cancer23, gastric cancer24, lung cancer25, and others26,27. CircRNA resists degradation by exonucleases, making it advantageous for clinical diagnosis, treatment, and prognosis of various diseases28. Consequently, our study concentrated on the expression patterns and mechanisms of circRNA in HCC. In this study, circ-0015004 was screened and validated from the GSE12174 dataset. We observed a remarkable upregulation in both HCC tissues and cell lines. A higher expression level of circ-0015004 was associated with a worse TNM stage and a larger tumor size. Then, we confirmed its resistance to RNase R, indicating it possesses the characteristics of circRNA. We also found that it is primarily localized in the cytoplasm. To illustrate circRNA's role in the malignant transformation of HCC, we conducted a series of functional experiments in vitro and vivo. Knocking down circ-0015004 expression suppressed the proliferation of HCC cells. Furthermore, we demonstrated that circ0015004 acts as a sponge for miR-330-3p, thereby inhibiting its function. Since miR-330-3p suppresses the translation of RCC2, we elucidated a novel signaling pathway, the circ0015004/miR-330-3p/RCC2 axis, in promoting HCC growth. Finally, we discovered that YY1 binding at the SDHC promoter region promotes circ-0015004 expression.
It is well documented that circRNAs play as miRNAs sponges to regulate gene expression and cellular activities. Our study unraveled the role of circ-0015004 as an antagonistic RNA (ceRNA) countering miR-330-3p, subsequently enhancing the mRNA level of RCC2 in HCC. MiR-330-3p has been acknowledged for its suppressive effects on various human tumors, including colorectal cancer, melanoma, and ovarian cancer29,30,31. Notably, miR-330-3p downregulation amplifies HCC cell proliferation32,33. In line with the previous studies, miR-330-3p expression was decreased in both HCC cell lines and tissues. Moreover, the increased miR-330-3p significantly restricted the growth of HCC cells, underscoring the potential function of circ-0015004 as a modulator of miR-330-3p in HCC.
To further explore the mechanism by which circRNA exerts its biological functions through miRNA sponges, we utilized three databases to predict downstream targets. We identified RCC2, which has been previously studied and shown to play a pivotal role in regulating RAC1 activity. RCC2 is of great interest because it is involved in multiple cellular mechanisms, including cell proliferation, invasion, migration, apoptosis, cell cycle progression, and DNA damage response34,35,36. Noteworthy, RCC2 expression levels in HCC tissue correlate with tumor stage, size, differentiation and metastasis, influencing patients' median survival times. Moreover, ectopic RCC2 expression engenders an escalation in colony formation and HCC cell proliferation while influencing cellular apoptosis37,38. Our study highlighted an elevation in RCC2 expression within both HCC tissues and cell lines. Importantly, we demonstrated that circ-0015004 could upregulate the expression of RCC2 by sponging miR-330-3p.
To further investigate the generation mechanism of circ-0015004, we utilized JASPAR to predict the upstream transcription factors of parent gene. We identified YY1 as a potential upstream transcription factor. YY1 is a transcription factor that is elevated in cancer cells and regulates genes involved in differentiation, cell cycle control and carcinogenesis39,40,41. Its dysregulation is associated with a variety of cancers, and its expression is increased in HCC42. YY1 promotes quaking expression via super-enhancer binding during EMT of HCC40. Our study further demonstrated that YY1 upregulates circ-0015004 expression, consequently impacting miR-330-3p and RCC2 mRNA levels.
The unique biological structure and functions of circRNAs present opportunities for early diagnosis, clinical treatment, and prognosis monitoring in patients with HCC. The YY1-mediated upregulation of circ-0015004 modulates the expression of RCC2 by regulating the function of miR-330-3p, elucidating the specific mechanism underlying the biological functions of circ-0015004. Validation of downregulating circ-0015004 expression to suppress tumor proliferation both in vitro and in vivo underscores the potential therapeutic targeting of circ-0015004 for HCC. However, our research primarily consisted of in vitro studies and did not extend to more profound animal experiments, such as knockout mice, or clinical trials. Consequently, the potential clinical application of circ-0015004 remains uncertain, highlighting the necessity for further exploration in future studies.
Conclusions
In conclusion, we have identified a novel circular RNA, circ0015004, which exhibits upregulation in HCC. The expression of circ0015004, under the regulation of YY1, correlates positively with the grade of HCC. Our functional analyses have demonstrated that circ0015004 facilitates progression of HCC, both in vitro and in vivo. Mechanistically, circ0015004 enhances the activity of RCC2, an established oncogenic protein, by sequestering mir-330-3p and thereby abrogating its inhibitory effect on RCC2 mRNA translation. Moreover, YY1 promotes the expression of circ0015004. These findings suggest that circ0015004 holds promise as a novel prognostic biomarker and potential therapeutic target for HCC (Fig. 8).
Data availability
The data of this study are available from the corresponding author upon reasonable request.
References
Yang, J. D. & Heimbach, J. K. New advances in the diagnosis and management of hepatocellular carcinoma. BMJ-Br. Med. J. 371, 15. https://doi.org/10.1136/bmj.m3544 (2020).
Siegel, R. L., Miller, K. D., Wagle, N. S. & Jemal, A. Cancer statistics, 2023. CA-Cancer J. Clin. 73, 17–48. https://doi.org/10.3322/caac.21763 (2023).
Morgan, E. et al. Global burden of colorectal cancer in 2020 and 2040: Incidence and mortality estimates from GLOBOCAN. Gut 72, 338–344. https://doi.org/10.1136/gutjnl-2022-327736 (2023).
Papatheodoridi, M., Tampaki, M., Lok, A. S. & Papatheodoridis, G. V. Risk of HBV reactivation during therapies for HCC: A systematic review. Hepatology 75, 1257–1274. https://doi.org/10.1002/hep.32241 (2022).
Liu, Z. Y. et al. Molecular targeted and immune checkpoint therapy for advanced hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 38, 13. https://doi.org/10.1186/s13046-019-1412-8 (2019).
Llovet, J. M. et al. Hepatocellular carcinoma. Nat. Rev. Dis. Primers 7, 28. https://doi.org/10.1038/s41572-020-00240-3 (2021).
Kristensen, L. S., Jakobsen, T., Hager, H. & Kjems, J. The emerging roles of circRNAs in cancer and oncology. Nat. Rev. Clin. Oncol. 19, 188–206. https://doi.org/10.1038/s41571-021-00585-y (2022).
Xu, J. J. et al. CircRNA-SORE mediates sorafenib resistance in hepatocellular carcinoma by stabilizing YBX1. Signal Transduct. Target. Ther. 5, 14. https://doi.org/10.1038/s41392-020-00375-5 (2020).
Liu, L. Y. et al. CircGPR137B/miR-4739/FTO feedback loop suppresses tumorigenesis and metastasis of hepatocellular carcinoma. Mol. Cancer 21, 17. https://doi.org/10.1186/s12943-022-01619-4 (2022).
Du, A. S. et al. M6A-mediated upregulation of circMDK promotes tumorigenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. Mol. Cancer 21, 18. https://doi.org/10.1186/s12943-022-01575-z (2022).
Liu, W. W. et al. Circ-ZEB1 promotes PIK3CA expression by silencing miR-199a-3p and affects the proliferation and apoptosis of hepatocellular carcinoma. Mol. Cancer 21, 15. https://doi.org/10.1186/s12943-022-01529-5 (2022).
Li, Y. et al. Hsa_circ_0000098 is a novel therapeutic target that promotes hepatocellular carcinoma development and resistance to doxorubicin. J. Exp. Clin. Cancer Res. 41, 17. https://doi.org/10.1186/s13046-022-02482-3 (2022).
Cen, J. J. et al. Circular RNA circSDHC serves as a sponge for miR-127-3p to promote the proliferation and metastasis of renal cell carcinoma via the CDKN3/E2F1 axis. Mol. Cancer 20, 14. https://doi.org/10.1186/s12943-021-01314-w (2021).
Rupaimoole, R. & Slack, F. J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 16, 203–221. https://doi.org/10.1038/nrd.2016.246 (2017).
Yi, J. et al. CircPVT1 promotes ER-positive breast tumorigenesis and drug resistance by targeting ESR1 and MAVS. EMBO J. 42, 20. https://doi.org/10.15252/embj.2022112408 (2023).
Li, J. et al. CircRPN2 inhibits aerobic glycolysis and metastasis in hepatocellular carcinoma. Cancer Res. 82, 15. https://doi.org/10.1158/0008-5472.Can-21-1259 (2022).
Huang, G. Q. et al. CircRNA hsa_circRNA_104348 promotes hepatocellular carcinoma progression through modulating miR-187-3p/RTKN2 axis and activating Wnt/β-catenin pathway. Cell Death Dis. 11, 14. https://doi.org/10.1038/s41419-020-03276-1 (2020).
Diener, C., Keller, A. & Meese, E. Emerging concepts of miRNA therapeutics: From cells to clinic. Trends Genet. 38, 613–626. https://doi.org/10.1016/j.tig.2022.02.006 (2022).
Liu, L. H., Tian, Q. Q., Liu, J., Zhou, Y. & Yong, H. M. Upregulation of hsa_circ_0136666 contributes to breast cancer progression by sponging miR-1299 and targeting CDK6. J. Cell. Biochem. 120, 12684–12693. https://doi.org/10.1002/jcb.28536 (2019).
Yang, J. H. et al. CircRNA_09505 aggravates inflammation and joint damage in collagen-induced arthritis mice via miR-6089/AKT1/NF-κB axis. Cell Death Dis. 11, 13. https://doi.org/10.1038/s41419-020-03038-z (2020).
Zhao, Q. Y. et al. Targeting mitochondria-located circRNA SCAR alleviates NASH via reducing mROS output. Cell 183, 76. https://doi.org/10.1016/j.cell.2020.08.009 (2020).
Luo, Z. J. et al. CircCAMSAP1 promotes hepatocellular carcinoma progression through miR-1294/GRAMD1A pathway. J. Cell. Mol. Med. 25, 3793–3802. https://doi.org/10.1111/jcmm.16254 (2021).
Wu, D. J., Li, Y. Q., Xu, A. J., Tang, W. Q. & Yu, B. CircRNA RNA hsa_circ_0008234 promotes colon cancer progression by regulating the miR-338-3p/ETS1 axis and PI3K/AKT/mTOR signaling. Cancers 15, 16. https://doi.org/10.3390/cancers15072068 (2023).
Zhang, X. et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol. Cancer 18, 24. https://doi.org/10.1186/s12943-018-0935-5 (2019).
Cui, Y. M. et al. CircHERC1 promotes non-small cell lung cancer cell progression by sequestering FOXO1 in the cytoplasm and regulating the miR-142-3p-HMGB1 axis. Mol. Cancer 22, 20. https://doi.org/10.1186/s12943-023-01888-7 (2023).
Liu, Z. H. et al. Circular RNA hsa_circ_001783 regulates breast cancer progression via sponging miR-200c-3p. Cell Death Dis. 10, 14. https://doi.org/10.1038/s41419-018-1287-1 (2019).
Gu, L. A. et al. circCYP24A1 facilitates esophageal squamous cell carcinoma progression through binding PKM2 to regulate NF-κB-induced CCL5 secretion. Mol. Cancer 21, 17. https://doi.org/10.1186/s12943-022-01686-7 (2022).
Kristensen, L. S. et al. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 20, 675–691. https://doi.org/10.1038/s41576-019-0158-7 (2019).
Louis, C., Leclerc, D. & Coulouarn, C. Emerging roles of circular RNAs in liver cancer. JHEP Rep. 4, 16. https://doi.org/10.1016/j.jhepr.2021.100413 (2022).
Wang, M., Yu, F., Chen, X. Z., Li, P. F. & Wang, K. The underlying mechanisms of noncoding RNAs in the chemoresistance of hepatocellular carcinoma. Mol. Ther. Nucleic Acids 21, 13–27. https://doi.org/10.1016/j.omtn.2020.05.011 (2020).
Qiu, L. P. et al. Circular RNAs in hepatocellular carcinoma: Biomarkers, functions and mechanisms. Life Sci. 231, 11. https://doi.org/10.1016/j.lfs.2019.116660 (2019).
Huang, Y. G. et al. HLA-F-AS1/miR-330-3p/PFN1 axis promotes colorectal cancer progression. Life Sci. 254, 9. https://doi.org/10.1016/j.lfs.2019.117180 (2020).
Yao, Y., Zuo, J. & Wei, Y. G. Targeting of TRX2 by miR-330-3p in melanoma inhibits proliferation. Biomed. Pharmacother. 107, 1020–1029. https://doi.org/10.1016/j.biopha.2018.08.058 (2018).
Yu, H. et al. RCC2 promotes proliferation and radio-resistance in glioblastoma via activating transcription of DNMT1. Biochem. Biophys. Res. Commun. 516, 999–1006. https://doi.org/10.1016/j.bbrc.2019.06.097 (2019).
Bruun, J. et al. Regulator of chromosome condensation 2 identifies high-risk patients within both major phenotypes of colorectal cancer. Clin. Cancer Res. 21, 3759–3770. https://doi.org/10.1158/1078-0432.Ccr-14-3294 (2015).
Pang, B. et al. Overexpression of RCC2 enhances cell motility and promotes tumor metastasis in lung adenocarcinoma by inducing epithelial-mesenchymal transition. Clin. Cancer Res. 23, 5598–5610. https://doi.org/10.1158/1078-0432.Ccr-16-2909 (2017).
Chen, Q. M., Jiang, P. Q., Jia, B. X., Liu, Y. H. & Zhang, Z. RCC2 contributes to tumor invasion and chemoresistance to cisplatin in hepatocellular carcinoma. Hum. Cell 33, 709–720. https://doi.org/10.1007/s13577-020-00353-7 (2020).
Xiong, D. D. et al. High throughput circRNA sequencing analysis reveals novel insights into the mechanism of nitidine chloride against hepatocellular carcinoma. Cell Death Dis. 10, 16. https://doi.org/10.1038/s41419-019-1890-9 (2019).
Cunningham, J. T. et al. mTOR controls mitochondrial oxidative function through a YY1-PGC-1α transcriptional complex. Nature 450, 736-U712. https://doi.org/10.1038/nature06322 (2007).
Han, J. X. et al. YY1 complex promotes quaking expression via super-enhancer binding during EMT of hepatocellular carcinoma. Cancer Res. 79, 1451–1464. https://doi.org/10.1158/0008-5472.Can-18-2238 (2019).
Wang, J. et al. YY1 positively regulates transcription by targeting promoters and super-enhancers through the BAF complex in embryonic stem cells. Stem Cell Rep. 10, 1324–1339. https://doi.org/10.1016/j.stemcr.2018.02.004 (2018).
Yang, W. D. et al. YY1 promotes endothelial cell-dependent tumor angiogenesis in hepatocellular carcinoma by transcriptionally activating VEGFA. Front. Oncol. 9, 12. https://doi.org/10.3389/fonc.2019.01187 (2019).
Funding
This study was supported by the Changzhou Applied Basic Research Program's Medical Research Project (CJ20220231) and the Project of Nanjing Municipal Health Commission (ZKX17036).
Author information
Authors and Affiliations
Contributions
Jie Z and Tong Z conceptualized and formulated the experimental design. PW, KJW, JJQ and Jie Z conducted the experiments. LQS, Jun Z and Qiang Z scrutinized and analyzed the data. LQS, Jun Z and Qiang Z composed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhao, J., Zhang, T., Wu, P. et al. circRNA-0015004 act as a ceRNA to promote RCC2 expression in hepatocellular carcinoma. Sci Rep 14, 16913 (2024). https://doi.org/10.1038/s41598-024-67819-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-67819-8
- Springer Nature Limited