Abstract
Despite recent advances in surgical techniques and perinatal management in obstetrics for reducing intraoperative bleeding, blood transfusion may occur during a cesarean section (CS). This study aims to identify machine learning models with an optimal diagnostic performance for intraoperative transfusion prediction in parturients undergoing a CS. Additionally, to address model performance degradation due to data imbalance, this study further investigated the variation in predictive model performance depending on the ratio of event to non-event data (1:1, 1:2, 1:3, and 1:4 model datasets and raw data).The area under the receiver operating characteristic curve (AUROC) and area under the precision-recall curve (AUPRC) were evaluated to compare the predictive accuracy of different machine learning algorithms, including XGBoost, K-nearest neighbor, decision tree, support vector machine, multilayer perceptron, logistic regression, random forest, and deep neural network. We compared the predictive performance of eight prediction algorithms that were applied to five types of datasets. The intraoperative transfusion in maternal CS was 7.2% (1020/14,254). XGBoost showed the highest AUROC (0.8257) and AUPRC (0.4825) among the models. The most significant predictors for transfusion in maternal CS as per machine learning models were placenta previa totalis, haemoglobin, placenta previa partialis, and platelets. In all eight prediction algorithms, the change in predictive performance based on the AUROC and AUPRC according to the resampling ratio was insignificant. The XGBoost algorithm exhibited optimal performance for predicting intraoperative transfusion. Data balancing techniques employed to alter the event data composition ratio of the training data failed to improve the performance of the prediction model.
Similar content being viewed by others
Introduction
The massive bleeding associated with a cesarean section (CS) can lead to severe postoperative complications and mortality for parturients1,2. In particular, if placenta abnormalities are present, the amounts of blood loss and transfusion during surgery tend to be large and urgent, having a decisive impact on the parturient prognosis3. Peripartum transfusion rates have been reported to reach 0.2–3.2%4,5 and up to 68.8% for mothers at risk of massive bleeding, such as those with placenta previa6. Therefore, early detection of risk factors associated with bleeding and the prediction for the need of a transfusion are essential in managing a CS. While some risk factors for bleeding and transfusion during the maternal delivery cycle have been documented, the majority of attention has been directed towards postpartum hemorrhage7,8,9,10. There is currently a shortage of large-scale clinical studies dedicated to identifying risk factors and developing predictive models for intraoperative blood transfusion during CS. As the surgical technique and intraoperative and postoperative maternal patterns of CS may differ from those of vaginal birth, these aspects need to be discussed separately, and therefore, large-scale studies focusing on intraoperative blood transfusion during CS are needed. Such research is crucial for ensuring the effective allocation of transfusion resources during CS, proper preoperative preparation of peripheral or central venous lines, and adequate staffing of operating rooms.
Recent advances in computer science and technology have enabled the development of machine learning (ML), a type of artificial intelligence (AI), which offers superior predictive abilities over existing models11. Models trained through various ML algorithms have demonstrated an excellent diagnostic performance in predicting postoperative mortality, complications, and prognosis12,13. Some recent studies have used ML models to predict peripartum maternal bleeding and transfusion10,14,15,16,17. Using ML to predict maternal bleeding and the need for a transfusion, correlations not found in conventional linear statistical analysis can be observed. However, most studies using ML have primarily focused on postpartum hemorrhage associated with vaginal delivery. To date, studies on ML prediction models for CS are lacking, and the efficacy of predictive ML techniques for a blood transfusion during a CS remains unclear.
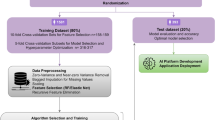
Therefore, in this study, we aimed to build ML models having the best diagnostic performances for predicting the need for an intraoperative red blood cell (RBC) transfusion during a CS and compare their suitability. The analysis used extreme gradient boost (XGBoost), K-nearest neighbor (KNN), decision tree (DT), support vector machine (SVM), multilayer perceptron (MLP), logistic regression (LR), random forest (RF), and deep neural network (DNN) algorithms. Furthermore, we attempted to identify an efficient approach to addressing a data imbalance by comparing the predictive performance of eight prediction algorithms applied to five types of datasets (1:1, 1:2, 1:3, and 1:4 model datasets and raw data).
Methods
Study design and parturients
This retrospective study was approved by the Institutional Review Board of the Asan Medical Center (protocol number 2021-0812) and was conducted in accord with the Declaration of Helsinki. The need for written informed consent was waived. Parturients who underwent a CS from January 1, 2010 to December 31, 2020 were included. Parturients with incomplete data or missing laboratory values were excluded from this study.
Data collection and study outcomes
For the input features of predictive modeling, we incorporated preoperative laboratory test results, risk factors highlighted in previous studies, and perioperative variables as suggested by clinical experts. All parturient data, including demographic data, perioperative variables, and laboratory values on preoperative days, were collected from an electronic medical record system. The demographic data included age, weight, height, body mass index (BMI), parity numbers, gestational diabetes mellitus (DM), placenta previa totalis/partialis/marginalis, placenta accreta/increta/percreta, placental abruption, pre-eclampsia, twin, and triple pregnancy. Perioperative variables included the type of anesthesia, midazolam use and intraoperative RBC transfusion. Preoperative laboratory tests of the parturients consisted of the most recent data values taken in the ward within two days prior to surgery. Preoperative laboratory values included white blood cell (WBC) count, hemoglobin, platelet count, neutrophil percent, lymphocyte percent, red cell distribution width (RDW), international normalized ratio (INR), neutrophil to lymphocyte ratio (NLR), platelet to lymphocyte ratio (PLR), prognostic nutritional index (PNI), estimated glomerular filtration rate (eGFR), creatinine, uric acid, albumin, aspartate transaminase (AST), alanine aminotransferase (ALT), total bilirubin, sodium, potassium, and chloride. The NLR was determined based on the ratio between the absolute neutrophil count and absolute lymphocyte count. The PLR was determined based on the ratio between the absolute platelet count and absolute lymphocyte count. The PNI was calculated as 10 × serum albumin (g/dl) + 0.005 × total lymphocyte count (per mm3). This study focused exclusively on red blood cells, excluding other blood products such as platelets and fresh frozen plasma, which were the transfusion products we aimed to predict. The primary aim was to select the ML model with the best performance in predicting the need for an intraoperative RBC transfusion during a CS. The secondary aim was to compare the prediction performance by applying the eight prediction algorithms to the five datasets (1:1, 1:2, 1:3, and 1:4 model datasets and raw data). Additionally, to investigate the impact of different combinations of input variables, or feature combinations, on predictive performance, we constructed several training datasets based on these combinations. Then, the performance of models trained on these varied datasets was comparatively analyzed.
Analysis and preprocessing of the dataset
Of the 16,137 parturients who were initially enrolled in the study, 1,883 were excluded due to incomplete demographic data, including missing information on height, weight, and comorbidities (n = 962), as well as incomplete laboratory values (n = 921). The parturients excluded from the study due to missing laboratory values accounted for approximately 5% of the total participants. Hence, 14,254 parturients were enrolled in this study. The number of parturients who received a RBC transfusion during surgery was 1020, that is, 7.16%. A dataset for predictive modeling was constructed by sampling data from parturients who received and those who did not receive a RBC transfusion. Alternatively, data from 1020 parturients randomly extracted from among the 13,234 parturients who did not receive a RBC transfusion and data from 1020 parturients who received a RBC transfusion were combined to form an equivalent ratio dataset and used as the training dataset of the 1:1 model. Furthermore, data from multiple numbers of the 1020 out of 13,234 parturients who did not receive a RBC transfusion were extracted, and datasets for the 1:2, 1:3, and 1:4 models were respectively constructed using data from 1020 patients who received a RBC transfusion. We used the bootstrap method to address the selection bias that occurs when sampling nonevent data. The bootstrap method was used to robustly evaluate the performance of the model by repeating the resampling process of the training data and thereby addressing the data imbalance. In this study, the average performance of the individual models was evaluated by resampling the training data 50 times and learning the extracted data. The missing values were removed during the modeling because there were no special mechanisms in which missing data occurred and the correlation between the missing variables was low (Supplementary Figure S1). All continuous input variables used in the predictive modeling were standardized using the StandardScaler provided by the Scikit-learn package 18. Categorical variables were input into the model through one-hot encoding.
ML models
As algorithms for predictive modeling, ML techniques such as KNN, DT, MLP, SVM, and LR; tree-based ensemble algorithms such as RF and XGBoost; and a simple five-layer DNN were used19,20,21,22,23,24,25,26. The entire dataset was divided into training, validation, and test datasets at a ratio of 6:2:2 for creating predictive models using 8 ML algorithms. The hyperparameters of all algorithms were tuned using the grid search method to achieve the best predictive performance for each model (Supplementary Methods).
Predictive performances
The predictive performance of each algorithm was evaluated based on the area under the receiver operating characteristics curve (AUROC) and area under the precision-recall curve (AUPRC), and the predictive results achieved through multiple bootstrap implementations were expressed based on the means and confidence intervals. The AUROC and AUPRC of each model were statistically and numerically compared. The predictive performances of the models when applying the resampling datasets were compared with that of the model using all raw data. Shapley additive explanation (SHAP) values were used to extract the feature importance of the predictive model used in this study. The SHAP value is a numerical expression of the influence on the direction and range of the contribution to the feature prediction27.
Statistical analysis
Continuous variables were expressed through the means and standard deviations, whereas categorical variables were expressed as numbers and percentages. Categorical data were analyzed using the Chi-square test or Fisher’s exact test, and continuous data were evaluated using an independent t-test or Mann–Whitney U test. Variables with two-tailed p-values of < 0.05 were considered statistically significant. ML modeling were conducted in Python 3.9 using the Scikit-Learn and TensorFlow packages.
Ethical approval
This retrospective research was approved by Institutional Review Board (IRB) of Asan Medical Center. Written informed consent was waived by the IRB (Asan Medical Center, No. 2021-0812).
Results
Table 1 shows the data characteristics of each group among the non-transfusion and transfusion receiving parturients. There were significant differences in weight (P < 0.001), height (P = 0.02), BMI (P = 0.004), parity numbers (P < 0.001), placenta previa totalis/partialis/marginalis (P < 0.001), placenta accreta/increta/percreta (P < 0.001), placental abruption (P < 0.001), twin pregnancy, (P = 0.003), triple pregnancy (P < 0.001), and postoperative RBC transfusion (P < 0.001) between the two groups. Regarding laboratory variables, there were significant differences in hemoglobin (P < 0.001), platelet (P < 0.001), RDW (P < 0.001), INR (P = 0.02), PLR (P = 0.02), PNI (P < 0.001), and estimated glomerular filtration rate (P < 0.001). Given the absence of discernible patterns in the distribution of missing data, we opted to exclude missing values from the analysis (Supplementary Figure S2).
Comparing the predictive performance of models
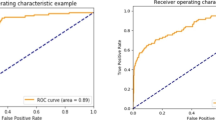
Table 2 shows the AUROC, AUPRC, accuracy, recall, precision, and F1 score of each ML model for an intraoperative RBC transfusion. Excluding the recall, the XGBoost model showed the best diagnostic performance based on the AUROC, AUPRC, accuracy, precision, and F1 score. When comparing the AUROC and AUPRC curves of each ML model for an intraoperative RBC transfusion. XGBoost achieved the highest AUROC (0.8257, 95% CI 0.8169–0.8345) and AUPRC (0.4825, 95% CI 0.4658–0.4992) (Fig. 1). The AUROC values of the KNN, DT, SVM, MLP, LR, RF, and DNN models were 0.5415 (95% CI 0.5336–0.5494), 0.7139 (95% CI 0.7040–0.7238), 0.6451 (95% CI, 0.6395–0.6507), 0.7509 (95% CI 0.7406–0.7612), 0.7382 (95% CI 0.7278–0.7486), 0.7756 (95% CI, 0.7744–0.7768), and 0.7719 (95% CI 0.7627–0.7811), respectively. The AUPRC values of the KNN, DT, SVM, MLP, LR, RF, and DNN models were 0.1155 (95% CI 0.1068–0.1242), 0.356 (95% CI 0.190–0.522), 0.2384 (95% CI, 0.2276–0.2492), 0.305 (95% CI 0.150–0.460), 0.2629 (95% CI 0.2492–0.2766), 0.4077 (95% CI, 0.3918–0.4236), and 0.4036 (95% CI 0.3900–0.4172), respectively. Figure 2 shows a comparison of the AUROC curves when applying the eight prediction algorithms to the five datasets (1:1, 1:2, 1:3, and 1:4 model datasets and raw data). For all eight prediction algorithms, the change in predictive performance based on the AUROC according to the resampling ratio was insignificant. Comparing the AUROC values of each prediction algorithm, XGBoost showed the best prediction performance (AUROC of 0.817–0.825). Figure 3 shows a comparison of the AUPRC curves when applying the eight prediction algorithms to the five datasets (1:1, 1:2, 1:3, and 1:4 model datasets and raw data). When each of the eight prediction algorithms was applied according to the resampling ratio, the predictive performances of the models learned using raw data based on the AUPRC were generally better than those of models learned using resampled datasets of other ratios. When comparing the AUPRC values of each prediction algorithm, XGBoost showed the best prediction performance compared with the AUPRC values (AUPRC of 0.429–0.478). When comparing the predictive performance of models based on various combinations of input variables, it was found that the combination including both patients' past medical histories and preoperative blood test findings demonstrated the highest performance, outperforming others in terms of both AUROC and AUPRC (Supplementary Figure S3).
Comparison of AUROC curves when applying eight prediction algorithms to five datasets (1:1, 1:2, 1:3, and 1:4 model datasets and raw data). (A) extreme gradient boost (XGBoost), (B) K-nearest neighbor (KNN), (C) decision tree (DT), (D) support vector machine (SVM), (E) multilayer perceptron (MLP), (F) logistic regression (LR), (G) random forest (RF), and (H) deep neural network (DNN).
Comparison of AUPRC curves when applying eight prediction algorithms to five datasets (1:1, 1:2, 1:3, and 1:4 model datasets and raw data). (A) extreme gradient boost (XGBoost), (B) K-nearest neighbor (KNN), (C) decision tree (DT), (D) support vector machine (SVM), (E) multilayer perceptron (MLP), (F) logistic regression (LR), (G) random forest (RF), and (H) deep neural network (DNN).
Feature importance
Figure 4 shows the feature importance of the XGBoost model when using the raw dataset. The feature importance of the XGBoost model shows that placenta previa totalis surgery, platelet level, preoperative hemoglobin level, and pre-eclampsia are important factors in predicting the need for an intraoperative RBC transfusion.
Discussion
In this study, we used an ML approach to compare models for predicting the necessity of an intraoperative RBC transfusion in parturients undergoing a CS. This study demonstrates that ML models can be useful for predicting an intraoperative RBC transfusion during a maternal CS, and among several ML models applied, the XGBoost algorithm showed the best predictive performance.
This study also shows how the performance of the predictive models varies according to the ratio of event data to nonevent data, providing important tips for the development of predictive models using medical data in which a data imbalance commonly occurs. In this study, training a model using a training dataset that matched an event-to-non-event ratio of 1:1 did not improve the predictive performance compared to using conventional raw data. Over-sampling and under-sampling methods can be applied in data-level approaches handling imbalanced data28,29. In particular, several over-sampling techniques have been introduced to strengthen the class boundaries, reduce an overfitting, and improve the discrimination. The most widely used technique is the synthetic minority over-sampling technique (SMOTE)28,29,30. Applying SMOTE to predictive modeling through ML is a commonly used approach to dealing with an imbalanced dataset28,29,30. Although it was initially believed that a 1:1 ratio of event versus non-event data in the training dataset would be an effective approach to training an imbalanced dataset, our findings suggest that artificially adjusting the event rate of a raw dataset does not lead to a better predictive performance of the model. Moreover, the predictive performances showed that models learned with raw data based on the AUPRC tended to be better than those learned with other balanced data. In this study, learning raw data with the original distribution rather than artificially changing the distribution exhibited a better overall predictive performance. These results are consistent with those shown in previous studies using large-scale electronic medical record (EMR) databases from multiple institutions13. Therefore, the performance of the prediction model cannot be improved by changing the event data composition ratio of the training data, as applied in this study. This imparts a critical lesson in predictive modeling using medical data. Data balancing techniques are unhelpful when modeling ML using an EMR dataset of numerical data.
In this study, the selection bias problems that arise when changing the event data ratios were addressed using the bootstrap method. The resampling process was repeated several times to prevent a degradation of the predictive performance depending on the specific data configuration.
In this study, XGBoost, a tree-based ensemble method, showed the best prediction performance in comparison to the other algorithms applied. In particular, it exhibited a better predictive performance than the DNN model. This is a different result from the conventional idea that a DNN model exhibits a superior predictive performance compared to other ML algorithms. These results were achieved because, as demonstrated in a previous study, tree-based ensemble methods such as XGBoost have shown a better predictive performance than DNN models when the number of input parameters of the model is relatively small13. Logistic regression is a commonly used traditional model in medical applications for the in-depth interpretation of clinical data. However, in this study, the logistic regression method showed a rather poor predictive power compared with other ML models. This indicates that machine learning approaches can achieve a better predictive power than recent classical statistical analysis. Despite extensive research on using existing statistical methods to develop prediction models for peripartum hemorrhage, limitations in predictive accuracy persist. Additionally, traditional models like decision trees and regression focused on fitting data closely, often leading to overfitting, whereas ML aims for broader generalization, doesn't assume specific data distributions unlike traditional regression, and better manages complex or large-scale data. Our findings advocate for the adoption of ML models as a more effective approach for such predictions.
Blood is a scarce and hard-to-obtain medical resource, and it is crucial not to waste it by preparing too much before surgery. Conversely, preparing too little blood can adversely affect the patient's outcome. Therefore, accurately predicting the amount of blood required before surgery is a critical aspect of clinical practice. In clinical practice, decisions regarding intraoperative blood transfusion primarily rely on either the actual volume of bleeding or predictions of massive bleeding based on the clinical expertise of anesthesiologists and obstetricians. If ML prediction models can be integrated into EMR systems in the future, maternal outcomes could be enhanced by facilitating tailored treatment strategies for the selection of high-risk parturients through the preparation of appropriate transfusion products and the setup of peripheral or central venous lines before CS. This integration could also lead to more efficient allocation of hospital resources, including optimizing the utilization of transfusion products and staffing operating rooms accordingly.
This study has certain limitations. First, this is a single-center study, and our results cannot be generalized to parturients undergoing a CS. Therefore, a multicenter study on this topic will be needed in the future. Additionally, this data are based on medical information composed from a single ethnic group. Further studies involving heterogeneous parturient groups of different races are needed. Second, we have not confirmed whether these predictive models contribute to improved clinical outcomes, such as reduced morbidity or mortality, in real-world settings. Future prospective studies are required to determine the impact of these models on patient outcomes when applied in actual clinical practice. Third, in this study, data balancing was only conducted at ratios of 1:1 to 1:4, and additional research results are therefore needed for further data balancing. In addition, more research is needed on the impact of data balancing techniques on predictions for outcomes with more variable event rates. Finally, the retrospective analysis might have been limited by missing clinical data, that is, important variables for intraoperative RBC transfusion prediction. Further prospective studies including such additional data are needed.
In conclusion, the predictive model using XGBoost showed the best predictive performance in predicting the need for an intraoperative RBC transfusion in parturients receiving a CS. In addition, in a study on ML prediction modeling using EMR datasets, similar to the current study, data balancing techniques that artificially control the event rates when constructing the training data did not improve the prediction performance. It will be necessary to evaluate through further research whether the findings of our study can be applied to larger datasets from multiple institutions.
Data availability
The dataset used and/or analyzed during the current study is available from the corresponding author on reasonable request. The codes that support the findings of this study are available for download on GitHub at https://github.com/SangWookLee20/CS_Transfusion. The dataset used and/or analyzed during the current study is available from the corresponding author on reasonable request. The codes that support the findings of this study are available for download on GitHub at https://github.com/SangWookLee20/CS_Transfusion.
References
Grobman, W. A. et al. Frequency of and factors associated with severe maternal morbidity. Obstet. Gynecol. 123, 804–810 (2014).
Haeri, S. & Dildy, G. A. 3rd. Maternal mortality from hemorrhage. Semin. Perinatol. 36, 48–55 (2012).
Lal, A. K. & Hibbard, J. U. Placenta previa: an outcome-based cohort study in a contemporary obstetric population. Arch. Gynecol. Obstet. 292, 299–305 (2015).
Mhyre, J. M. et al. Massive blood transfusion during hospitalization for delivery in New York State, 1998–2007. Obstet. Gynecol. 122, 1288–1294 (2013).
Van den Berg, K. et al. A cross-sectional study of peripartum blood transfusion in the Eastern Cape, South Africa. South Afr. Med. J. = Suid-Afrikaanse tydskrif vir geneeskunde 106, 1103–1109 (2016).
Spiegelman, J. et al. Risk factors for blood transfusion in patients undergoing high-order Cesarean delivery. Transfusion 57, 2752–2757 (2017).
Ehrenthal, D. B., Chichester, M. L., Cole, O. S. & Jiang, X. Maternal risk factors for peripartum transfusion. J. Women’s Health 2002(21), 792–797 (2012).
Ouh, Y.-T. et al. Predicting peripartum blood transfusion: focusing on pre-pregnancy characteristics. BMC Pregnancy Childbirth 19, 477 (2019).
Neary, C., Naheed, S., McLernon, D. J. & Black, M. Predicting risk of postpartum haemorrhage: A systematic review. Bjog 128, 46–53 (2021).
Ahmadzia, H. K., Phillips, J. M., James, A. H., Rice, M. M. & Amdur, R. L. Predicting peripartum blood transfusion in women undergoing cesarean delivery: A risk prediction model. PLoS One 13, e0208417 (2018).
Schmidt, J., Marques, M. R. G., Botti, S. & Marques, M. A. L. Recent advances and applications of machine learning in solid-state materials science. npj Comput. Mater. 5, 83 (2019).
Ren, Y. et al. Performance of a machine learning algorithm using electronic health record data to predict postoperative complications and report on a mobile platform. JAMA Netw. Open 5, e2211973 (2022).
Lee, S. W. et al. Multi-center validation of machine learning model for preoperative prediction of postoperative mortality. NPJ Digit Med 5, 91 (2022).
Akazawa, M., Hashimoto, K., Katsuhiko, N. & Kaname, Y. Machine learning approach for the prediction of postpartum hemorrhage in vaginal birth. Sci. Rep. 11, 22620 (2021).
Chen, H. et al. Construction and effect evaluation of prediction model for red blood cell transfusion requirement in cesarean section based on artificial intelligence. BMC Med Inform Decis Mak 23, 213 (2023).
Liu, J. et al. Machine learning-based prediction of postpartum hemorrhage after vaginal delivery: Combining bleeding high risk factors and uterine contraction curve. Arch Gynecol Obstet 306, 1015–1025 (2022).
Wang, Y., Xiao, J. & Hong, F. A risk prediction model of perinatal blood transfusion for patients who underwent cesarean section: A case control study. BMC Pregnancy Childbirth 22, 373 (2022).
Pedregosa, F. V. G. et al. Scikit-learn: Machine learning in python. J Mach Learn Res. 12, 2825–2830 (2011).
DR, C. The regression analysis of binary sequences. J. R. Stat. Soc. Ser. B (Methodological) 20(2), 215–242 (1958).
Breiman, L. Random forest. Mach. Learn. 45(1), 5–32 (2001).
LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521, 436–444 (2015).
Chen, T. Q. & Guestrin, C. XGBoost: A scalable tree boosting system. Kdd'16: Proceedings of the 22nd ACM Sigkdd international conference on knowledge discovery and data mining; https://doi.org/10.1145/2939672.2939785, 785–794 (2016).
Podgorelec, V., Kokol, P., Stiglic, B. & Rozman, I. Decision trees: an overview and their use in medicine. J Med Syst 26, 445–463 (2002).
Geva, S. & Sitte, J. Adaptive nearest neighbor pattern classification. IEEE Trans Neural Netw 2, 318–322 (1991).
Thomas, P. & Suhner, M. C. A new multilayer perceptron pruning algorithm for classification and regression applications. Neural Process Lett 42, 437–458 (2015).
Vapnik, V. N. The Nature of Statistical Learning Theory. 2nd edn, (Springer, 2000).
Lundberg, S. M., Lee, S. I. A unified approach to interpreting model predictions, 4765–4774 (2017).
Johnson, J. M. & Khoshgoftaar, T. M. Survey on deep learning with class imbalance. J. Big Data-Ger. 6 (2019).
Guo, H. X. et al. Learning from class-imbalanced data: Review of methods and applications. Expert Syst Appl 73, 220–239 (2017).
Chawla, N. V., Bowyer, K. W., Hall, L. O. & Kegelmeyer, W. P. SMOTE: Synthetic minority over-sampling technique. J Artif Intell Res 16, 321–357 (2002).
Acknowledgements
This research received no financial support from any funding agencies in the public, commercial, or not-for-profit sectors.
Funding
This research was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (Grant Number: RS-2022-00165755). This research was also supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (Grant Number: HR21C0198).
Author information
Authors and Affiliations
Contributions
Sang-Wook Lee: Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Software, Validation, Visualization, Writing – original draft. Bumwoo Park: Conceptualization, Data curation, Methodology, Resources, Supervision, Writing—review & editing. Jimung Seo: Data curation, Formal analysis. Sangho Lee: Data curation, Formal analysis. Ji-Hoon Sim: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing—original draft, Writing—review & editing. Sang-Wook Lee: Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Software, Validation, Visualization, Writing—original draft. Bumwoo Park: Conceptualization, Data curation, Methodology, Resources, Supervision, Writing—review & editing. Jimung Seo: Data curation, Formal analysis. Sangho Lee: Data curation, Formal analysis. Ji-Hoon Sim: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing—original draft, Writing—review & editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lee, SW., Park, B., Seo, J. et al. Development of a machine learning approach for prediction of red blood cell transfusion in patients undergoing Cesarean section at a single institution. Sci Rep 14, 16628 (2024). https://doi.org/10.1038/s41598-024-67784-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-67784-2
- Springer Nature Limited