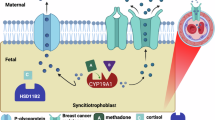
Abstract
Methadone maintenance treatment for opioid dependent mothers is standard of care. Infants of methadone maintained opioid dependent (MMOD) mothers have better outcomes compared to infants of opioid dependent mothers without treatment. However, when compared to non-exposed infants, infants of MMOD mothers are associated with worse outcomes. We conducted a pilot study to examine genome wide differential DNA methylation using cord blood samples from sixteen term and near-term infants of MMOD and opioid naïve mothers, excluding Infants with chorioamnionitis. A total of 152 differentially methylated loci were identified at a difference > + 2, < − 2 and p-value < 0.05. There were 90 hypermethylated loci (59 annotated genes) and 62 hypomethylated loci (38 annotated genes) observed. The hypermethylated and hypomethylated DNA changes involved multiple genes, pathways and networks that may explain some of the changes seen in infants of MMOD mothers. Top hypermethylated and hypomethylated genes involved areas of cell growth, neurodevelopment, vision and xenobiotic metabolism functions. Our data may explain the role of key pathways and genes relevant to neonatal outcomes seen from methadone exposure in pregnancy. Functional studies on the identified pathways and genes could lead to improved understanding of the mechanisms and identify areas for intervention.
Similar content being viewed by others
Introduction
Medication assisted treatment with methadone or buprenorphine is considered standard of care for opioid dependent pregnant persons1. When compared to untreated mothers using illicit opioids, methadone maintained opioid dependent (MMOD) mothers have better perinatal outcomes2. However, when compared to non-opioid exposed pregnant persons (or individuals), methadone exposure was associated with worse perinatal outcomes including increased risk for shorter gestation periods, lower birth weight, smaller head circumference, increased risk of developing neonatal opioid withdrawal syndrome (NOWS) and impaired visuocortical function at birth3,4,5. Also, children delivered to MMOD pregnant persons were found to exhibit lower psychomotor, cognitive, behavioral and language scores, in follow up studies6,7. Some proposed mechanisms by which opioid exposure influenced fetal development include inhibition of neuronal proliferation and differentiation with increased cell death, alterations in endocrine function and modifications to myelin sheath formations8,9.
DNA methylation is considered an important mechanism for changes seen in the developing fetus in response to medications and stress10. This idea is linked to the concept of Developmental Origins of Health and Disease (DOHad) hypothesis; a conceptual framework that links prenatal environmental exposures to subsequent health and disease outcomes later in life11. Epigenetic mechanisms, most importantly, changes in DNA methylation, has been suggested as one of the mechanisms of the outcomes seen in infants of MMOD mothers12. DNA methylation refers to the addition of a methyl (-CH3) group to the fifth position of cytosine nucleotide in areas of the genome where cytosine is followed immediately by guanine in the DNA sequence referred to as a CpG dinucleotide. This leads to the formation of 5-methylcytosine, an enzymatic 1-carbon metabolism process. Hypermethylated regions on genes are associated with decreased expression while hypomethylated regions are associated with increased gene expression13,14. Other epigenetic mechanisms like histone protein modification and non-coding RNAs have not been well studied as possible contributors to the associated outcomes.
Whole genome methylation profiling has been extensively utilized to evaluate the epigenetic basis of various human pathology15. Genome-wide DNA methylation profiling coupled with artificial intelligence (AI) has been used to create a pathway and network analysis with the aim of diagnosing maternal opioid exposure and predicting infants who will develop NOWS16. DNA methylation changes in opioid receptor related genes obtained from buccal samples of methadone-maintained mother and infant dyads found increased methylation in ABCB1, CYP2D6, and OPRM1 genes but no correlation with outcomes12.
To date, no study has examined differential DNA methylation regions (DMRs) in genome wide studies in umbilical cord blood (UCB) to form the basis of explaining neonatal outcomes for MMOD pregnant persons. Findings of DMRs in cord blood samples of infants of MMOD mothers may help explain some of the outcomes seen in these infants, especially if the methylation patterns persist and or correlate with gene expression associated with known disease processes. The aim of this pilot study is to determine if maternal methadone maintenance leads to differential neonatal DNA methylation patterns using UCB samples in full term and near-term infants.
We hypothesized that infants of MMOD persons will have differential DNA methylation compared to infants of opioid naïve persons.
Results
In this pilot study, we enrolled 16 infants (8 infants in the Methadone group and 8 infants in the Control group) and performed genome-wide DNA methylation on UCB DNA. At delivery all 16 infants appeared healthy. There were no significant differences in mean birth weight (2.76 ± 0.34 kg vs 3.12 ± 0.44, p = 0.06), mean gestational age (38.8 ± 0.9 vs 38.5 ± 1.5, p = 0.3), exposure to cigarette smoking (4/8 vs 2/8, p = 0.6) and other exposures, between the methadone exposed group and the control (Table 1).
Differential DNA methylation
DNA methylation levels are represented by β-values. The β-value is the ratio of the methylated probe signal intensity to the total locus intensity. The β-values range from 0 to 1 where 0 indicates unmethylated and 1 indicates fully methylated. A boxplot (Fig. 1) was generated from β-values of all probe sets from the 16 samples to describe distribution of the data. No significant differences were observed between the groups. A total of 152 differentially methylated loci were identified at a difference > + 2, < − 2 and p-value < 0.05, of which 90 are hypermethylated loci (59 annotated genes) and 62 are hypomethylated loci (38 annotated genes). The top 20 hyper and hypomethylated gene names and probe-IDs, in the exposure group, based on p-values are listed in Tables 2 and 3.
Cluster analysis: heatmap
Cluster analysis was performed on the 152 differentially methylated loci for the two sample groups using the Heatmap function in Partek Genomics Suite Software. A heatmap of the methylation levels for the 152 DNA methylation loci illustrates the differences between the two groups (Fig. 2).
Ingenuity pathway analysis
Ingenuity Pathway Analysis (IPA) software, Qiagen Inc., Germantown, MD) was used for pathway analysis by loading 152 probe sets (97 annotated genes) that were differentially methylated with exposure to methadone. A total of 325 canonical pathways were altered with exposure to methadone during pregnancy. Selected key pathways important in methadone pathophysiological response are shown in (Table 4, Figs. 3, and 4). Seventy-three diseases and functions were modified with methadone exposure. Selected key diseases and functions altered with methadone exposure are listed in Table 5.
Tox functions
Methadone exposure during pregnancy potentially altered 26 tox functions in UCB (Table 6). Tox functions related to cardiotoxicity include cardiac failure, cardiac arrhythmia, cardiac dysfunction, cardiac enlargement, and congenital heart anomaly. Altered hepatotoxic functions are hepatocellular cancer, liver hyperproliferation, liver inflammation/hepatitis, liver cirrhosis, liver fibrosis, and liver cholestasis. Altered tox functions related to nephrotoxicity are renal failure, glomerular injury, renal nephritis, renal inflammation, and renal cell necrosis/cell death.
Upstream regulators
Upstream regulator analysis by IPA identified 49 regulators to be dysregulated. Key identified regulators and their target molecules are described in Table 7.
Discussion
This pilot study found that there is differential DNA methylation in UCB cells from infants of MMOD persons involving multiple areas of body function. Exposure to methadone was associated with DNA methylation changes in genes that can contribute to neurological, neurobehavioral and growth abnormalities. Prior studies have observed increased risk of prematurity, smaller body measurements, increased susceptibility to opioid withdrawal and poorer neurodevelopmental outcomes with prenatal exposure to methadone3,4,5,6,7. In addition, Kelty, et al., showed association with increased risk of certain immune-related conditions like asthma and eczema in children with prenatal opioids exposure17. Although the mechanisms of these findings are not yet known, alteration in gene expression secondary to differential DNA methylation may potentially contribute to this increased risk.
The top hypermethylated genes observed with methadone exposure include CPLX4 (complexin 4), ZFP3 (ZFP3 Zinc Finger Protein), PLEKHA7 (pleckstrin homology domain containing A7), KCNC1 (potassium voltage-gated channel subfamily C member 1), TANC1 (Tetratricopeptide Repeat, Ankyrin Repeat and Coiled-Coil Containing 1) and AUTS2 (Activator of Transcription and Developmental Regulator AUTS2). CPLX4 is associated with encoded protein involved in synaptic vesicle exocytosis. It has been linked to X- linked cone rod dystrophy which manifests as reduced visual acuity and sensitivity in the central visual field, leading to eventual peripheral vision loss and severe impairment of overall vision18. As mentioned above, some studies have associated prenatal methadone exposure to visual abnormalities that has not yet been well explained. Identifying the effect of hypermethylated CPLX4 on gene expression will be an important next step. ZFP3 is a protein coding gene that is suggested to enable DNA-binding transcription factor activity, RNA polymerase II-specific and RNA polymerase II transcription regulatory region sequence-specific DNA binding activity19. PLEKHA7, enables delta-catenin binding activity which is involved in epithelial cell–cell adhesion; pore complex assembly; and zonula adherens maintenance. It is found in several cellular components, including the centrosome; nucleoplasm; and zonula adherens. Diseases associated with PLEKHA7 include cleft lip with or without cleft palate and primary angle-closure glaucoma20. KCNC1 (potassium voltage-gated channel subfamily C member 1), plays an important role in the rapid repolarization of fast-firing brain neurons. Among its related pathways are potassium channels and transmission across chemical synapses21. TANC1 (Tetratricopeptide Repeat, Ankyrin Repeat and Coiled-Coil Containing 1) is predicted to be involved in the regulation of post synapse organization. TANC1 is associated with intellectual developmental disorders22. Likewise, AUTS2 (Activator of Transcription and Developmental Regulator AUTS2) has been implicated in neurodevelopment and identified as a candidate gene for numerous neurological disorders, including autism spectrum disorders, intellectual disability, and developmental delay23. Notably, neurodevelopmental delay has been reported in infants of MMOD mothers. Investigating alterations in the expression of these genes affected by DNA methylation and establishing correlations with neurodevelopmental outcomes represents a crucial next step in understanding the observed association. Top Hypomethylated genes include CLDN4 (claudin 4), TNIK (TRAF2 and NCK Interacting Kinase), ATP11A (ATPase Phospholipid Transporting 11A), BMP7 (Bone Morphogenetic Protein 7), FMN2 (Formin 2), and SCN5A (Sodium Voltage-Gated Channel Alpha Subunit 5). CLDN4 functions as a crucial membrane protein in the composition of epithelial cell tight junctions, regulating movement of solutes and ions through the paracellular space. CLDN4 is also believed to play a possible role in internal organ development and function during pre- and postnatal life. Its absence has been associated with Williams-Beuren syndrome, a neurodevelopmental disorder affecting multiple systems24. TNIK expression plays important roles in carcinogenesis and embryonic development. A mutation in this gene is associated with intellectual developmental disorder, characterized by significantly below-average general intellectual functioning accompanied by impairments in adaptive behavior, typically observed during the developmental period. Individuals with this disorder often exhibit intellectual disability, delayed speech, and hyperactivity25. ATP11A, which is integral to membrane ATPase function, has been linked with leukodystrophy, hypomyelinating diseases and deafness26. Abnormal myelination has been reported to infants of MMOD mothers27. BMP7 codes a ligand of growth factor of the TGF-beta superfamily that plays important role in various biological processes, including embryogenesis, hematopoiesis, neurogenesis, and skeletal morphogenesis28. Diseases associated with BMP7 include multiple types of congenital heart defects29. FMN2 encoded protein is thought to have essential roles in organization of the actin cytoskeleton and in cell polarity. Mutations in this gene have been associated with infertility and with an autosomal recessive form of intellectual disability30.
SCN5A protein mediates the voltage-dependent sodium ion permeability of excitable membranes. It forms a sodium-selective channel through which Na (+) ions may pass in accordance with their electrochemical gradient. Over expression may impair the function of excitable membranes31. Diseases associated with SCN5A include sudden infant death syndrome and long QT syndrome. Methadone use in adults can lead to prolongation of QTc through inhibition of the cardiac ion channel KCNH2 in a dose dependent manner32. In infants of MMOD mothers, there have also been reports of QTc prolongation in the first 2 days of life with subsequent normalization33. The mechanism of neonatal QTc prolongation related to MMOD has not been described. While we may assume it mirrors what was described in adults, it may also be related to epigenetic changes in certain excitable membranes like the observations made in the SCN5A gene.
Altered methylation pattern in genes involved in canonical pathways can potentially link methadone exposure during pregnancy to short- and long-term outcomes in offspring. At least 325 canonical pathways were found to be altered in UCB cells exposed to methadone during pregnancy, in this study. OPRM1, a mu-opioid receptor related gene, has been shown to be differentially methylated with methadone exposure and may play a role in NOWS risk or substance dependence later in life34. While this pilot study did not observe increased methylation with the OPRM1 gene, our data indicate that xenobiotic metabolism PXR signaling pathway and xenobiotic metabolism CAR signaling pathway were altered in infants of MMOD mothers. This change could imply an adaptive response to opioid exposure in the same manner as of increased methylation of OPRM1 with resultant varied response to opioid exposure later in life.
Moreover, we found that intrauterine exposure to methadone was associated with increased DNA methylation in genes related to the NRF2-mediated oxidative stress response pathway and superoxide radicals’ degradation pathways. Leventelis C, et al. and another study reported significant oxidative stress response with associated evidence of compromised antioxidant defense in adults exposed to heroin who were maintained on methadone35,36. Altered oxidative stress response in infants of MMOD may be reflective of in utero adaptation to methadone exposure. Oxidative stress has been shown to be detrimental to neuronal development. Obst S, et al. found that increased reactive oxidative species (ROS) have detrimental effects on oligodendrocyte maturation, myelination, and neuronal survival, leading to ultrastructural abnormalities of myelin formation and grey matter injury37. Abnormal oligodendrocyte maturation and altered maturation of connective tracts have been associated with infants of MMOD mothers with suggestion that this finding may be the basis of the increased risk for cognitive and behavioral difficulties observed in children of mothers using opioids.27,38.
Altered intracellular growth hormone (GH) signaling pathways involving PIK3CG and SOCS7 molecules can affect the synthesis of insulin-like growth factors (IGF) associated with increased risk of fetal growth restriction observed in infants of MMOD mothers. Intrauterine human growth requires the normal expression of IGF-I/IGF-II and type 1-IGF receptor39. While IGF-receptor genes have not been implicated in the canonical pathway, IGF-I and IGF-II expression can be impaired secondary to upstream cytoplasmic alteration in growth hormone signaling. Fetal growth in the third trimester is mostly influenced by nutritional and other maternal factors and less by fetal genetics. Nonetheless, persistent abnormalities in the GH-IGF axis have been implicated in small for gestational age infants and fetal growth restriction40.
Exposure to methadone during pregnancy was associated with alterations in 26 toxic functions in our cohort. The top toxic functions that altered DNA methylation were seen in genes related to hepatoxicity, cardiotoxicity, and nephrotoxicity. The mechanism of cardiac rhythm abnormalities in infants of MMOD is not well described and epigenetic factors may play a contributory role.
To our knowledge, this is the first study reporting genome wide differential DNA methylation in UCB cells from infants exposed to methadone during pregnancy. The altered methylation pattern identified in genes relate to neurodevelopmental delay, neurobehavioral disorders, oxidative stress and growth function show a potential connection to the reduced neurodevelopmental and anthropometric measures observed in infants of MMOD mothers. An important follow up study is investigating how differential methylation influences gene expression in these cohorts of infants. Further studies, preferably with a larger cohort, is needed to investigate a cumulative dose-dependent effect of methadone on DNA methylation patterns and if observed changes in DNA methylation persist through out the neonatal period and if it correlates with certain outcomes seen in infants of MMOD mothers.
Our study has its limitations, particularly, the small sample size of 16 neonates, but similar limited sample sizes have been used in studies investigating differential DNA methylation patterns41,42. There is a chance of finding differences in DNA methylation due to multiple comparisons which was controlled by employing a FDR p-value < 0.05, and a beta difference of > + 2 or < − 2. Our results should be considered as hypothesis generating and should be validated in a larger cohort. Also, other maternal exposures including other substances used and maternal psychosocial stressors during pregnancy may contribute to the methylation changes observed.
In conclusion, methadone exposure during pregnancy is associated with differential DNA methylation in UCB cells. We identified 97 differentially methylated genes, important tox functions, upstream regulators and canonical pathways related to the oxidative stress, xenobiotic metabolic response, and cardiotoxicity in UCB cells of infants of MMOD mothers. Future studies can further validate differential methylation of target genes in a larger cohort of infants. Our data contribute to a deeper understanding of the impact of methadone exposure during pregnancy on both short-term and long-term outcomes, highlighting the significance of key pathways and genes. Functional studies on these identified pathways and genes could enhance our understanding of the underlying mechanisms, ultimately guiding the development of effective interventions.
Methods
Ethical, human study protocol and institutional biosafety approvals
All human protocols and procedures described in this study were approved by the Institutional Review Board of Thomas Jefferson University Hospital. All experiments performed in this study were approved by the Nemours Institutional Biosafety Committee. All methods were performed in accordance with the relevant guidelines and regulations. The Institutional Review Board (IRB) waived informed consent as the study was performed on discarded blood and placental tissue samples.
Study design
This is a pilot prospective observational study to examine differential DNA methylation in UCB cells of term and near-term neonates born to MMOD mothers. Samples of UCB and placental tissue were collected at the time of delivery from full term and near term (≥ 35 weeks) infants. Exclusion criteria included infants with fetal growth restriction, clinical or histological chorioamnionitis and major congenital/chromosomal anomalies.
UCB collection
The UCB was obtained on the delivery table immediately after separation of baby from placenta. When this method was not feasible, a long segment of the cord was obtained and cleaned with 70% alcohol prior to collection of samples using a needle and syringe or a butterfly needle. The UCB was collected in PAXGene blood DNA tube (BD Catalog # 7611650), processed on the day of collection as per manufacturer’s protocol and saved at – 80 °C.
Fetal membrane collection, processing, staining, and diagnosis of hierarchical cluster analysis (HCA)
The placental tissue was processed and kept in 10% neutral buffered formalin (NBF) for 24–48 h before it was transferred to 70% alcohol. Tissue samples were processed using standard operating procedures at the histopathology laboratory at Thomas Jefferson University and paraffin embedded in Histoplast LP (Thermo Fisher Scientific, Fremont, CA). The samples were then classified as having HCA or no HCA by a blinded pathologist (JC). We collected information on HCA as it is a potential confounder in assessing for DNA methylation based on a prior study showing that epigenetic changes occur via DNA methylation in infants exposed to HCA43,44.
DNA isolation and statistical approaches for DNA methylation analysis
DNA isolation was performed using QIAamp DNA Mini kit (Qiagen, Germantown, MD). DNA was quantified on a Qubit 2.0 fluorometer, (Thermo Fisher Scientific, Waltham, MA), and the DNA quality was assessed by an Agilent 2200 TapeStation (Agilent Technologies, Palo Alto, CA). The genome-wide DNA methylation study was performed using the Illumina Methylation EPIC Array (cat# WG-317-1001, Illumina Inc., San Diego, CA). Illumina iScan Reader was used to analyze the image and data from Methylation EPIC Bead Chip. Data processing was performed with Illumina GenomeStudio software. Raw IDAT files were processed using Partek Genomics Suite V.7.20 (Partek Inc. Missouri, USA) and annotated using the MethylationEPIC_v-1-0_B4 manifest file. Probes from the X and Y chromosomes were excluded from the study (since we are having both males and females in the samples), and probes based on detection with P > 0.05 were also filtered to exclude low-quality probes. Background normalization was performed using Swan Normalization. Principal component analysis (PCA) was performed to visualize clusters in the methylation data, and as a quality control procedure (Fig. 5). Distribution of β-values across the samples was inspected by a box-and-whiskers plot (Fig. 1). Samples were attributed to the two groups, methadone exposed and control. Differential methylation analysis was then performed between the two groups at a p-value < 0.05, and a difference of > + 2 or < − 2. To detect the differential methylation in global CpGs that varies across all samples, we performed a 1-way ANOVA test. Hierarchical cluster analysis of the significant CpGs was carried out with the Heatmap function in the Partek Genomics Suite (Fig. 2). Clinical data were compared with Fisher’s exact test and Wilcoxon rank-sum test using Stata Statistical Software 15 College Station, TX: Stata Corp LLC.
Data availability
The data that support the findings of this study are not openly available due to reasons of sensitivity and are available from the corresponding author upon reasonable request. Data are in an encrypted server with Thomas Jefferson University.
References
Mattick, R. P., Breen, C., Kimber, J. & Davoli, M. Buprenorphine maintenance versus placebo or methadone maintenance for opioid dependence. Cochrane Database Syst. Rev. 2014, CD02207 (2014).
Mattick, R. P., Breen, C., Kimber, J. & Davoli, M. Methadone maintenance therapy versus no opioid replacement therapy for opioid dependence. Cochrane Database Syst. Rev. 2009, CD002209 (2009).
Dryden, C., Young, D., Hepburn, M. & Mactier, H. Maternal methadone use in pregnancy: Factors associated with the development of neonatal abstinence syndrome and implications for healthcare resources. BJOG 116, 665–671 (2009).
Cleary, B. J. et al. Methadone and perinatal outcomes: A retrospective cohort study. Am. J. Obstet. Gynecol. 204(139), e1-9 (2011).
McGlone, L. et al. Neonatal visual evoked potentials in infants born to mothers prescribed methadone. Pediatrics 131, e857–e863 (2013).
Levine, T. A., Davie-Gray, A., Kim, H. M., Lee, S. J. & Woodward, L. J. Prenatal methadone exposure and child developmental outcomes in 2-year-old children. Dev. Med. Child Neurol. 63, 1114–1122 (2021).
Andersen, J. M., Høiseth, G. & Nygaard, E. Prenatal exposure to methadone or buprenorphine and long-term outcomes: A meta-analysis. Early Hum. Dev. 143, 104997 (2020).
Konijnenberg, C. & Melinder, A. Neurodevelopmental investigation of the mirror neurone system in children of women receiving opioid maintenance therapy during pregnancy. Addiction 108, 154–160 (2013).
Farid, W. O., Dunlop, S. A., Tait, R. J. & Hulse, G. K. The effects of maternally administered methadone, buprenorphine and naltrexone on offspring: Review of human and animal data. Curr. Neuropharmacol. 6, 125–150 (2008).
Godfrey, K. M., Lillycrop, K. A., Burdge, G. C., Gluckman, P. D. & Hanson, M. A. Epigenetic mechanisms and the mismatch concept of the developmental origins of health and disease. Pediatr. Res. 61, 5R-10R (2007).
Barker, D. J. P. The origins of the developmental origins theory. J. Intern. Med. 261, 412–417 (2007).
McLaughlin, P. et al. Increased DNA methylation of ABCB1, CYP2D6, and OPRM1 genes in newborn infants of methadone-maintained opioid-dependent mothers. J. Pediatr. 190, 180-184.e1 (2017).
Razin, A. & Riggs, A. D. DNA methylation and gene function. Science 210, 604–610 (1980).
Morley, R., Saffery, R., Hacking, D. F. & Craig, J. M. Epigenetics and neonatology: The birth of a new Era. Neoreviews 10, e387–e395 (2009).
Bahado-Singh, R., Vishweswaraiah, S., Mishra, N. K., Guda, C. & Radhakrishna, U. Placental DNA methylation changes in detection of tetralogy of Fallot. Ultrasound Obstet. Gynecol. 55, 768–775 (2020).
Radhakrishna, U. et al. Placental DNA methylation profiles in opioid-exposed pregnancies and associations with the neonatal opioid withdrawal syndrome. Genomics 113, 1127–1135 (2021).
Kelty, E., Rae, K., Jantzie, L. L., Wyrwoll, C. S. & Preen, D. B. Prenatal opioid exposure and immune-related conditions in children. JAMA Netw Open 7, e2351933 (2024).
Mäntyjärvi, M. et al. Clinical features and a follow-up study in a family with X-linked progressive cone-rod dystrophy. Acta Ophthalmol. Scand. 79, 359–365 (2001).
Fagerberg, L. et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteomics 13, 397–406 (2014).
Meng, W., Mushika, Y., Ichii, T. & Takeichi, M. Anchorage of microtubule minus ends to adherens junctions regulates epithelial cell-cell contacts. Cell 135, 948–959 (2008).
Muona, M. et al. A recurrent de novo mutation in KCNC1 causes progressive myoclonus epilepsy. Nat. Genet. 47, 39–46 (2015).
Kong, Y., Zhou, W. & Sun, Z. Nuclear receptor corepressors in intellectual disability and autism. Mol. Psychiatry 25, 2220–2236 (2020).
Beunders, G. et al. Two male adults with pathogenic AUTS2 variants, including a two-base pair deletion, further delineate the AUTS2 syndrome. Eur. J. Hum. Genet. 23, 803–807 (2015).
Paperna, T., Peoples, R., Wang, Y. K., Kaplan, P. & Francke, U. Genes for the CPE receptor (CPETR1) and the human homolog of RVP1 (CPETR2) are localized within the Williams-Beuren syndrome deletion. Genomics 54, 453–459 (1998).
Anazi, S. et al. A null mutation in TNIK defines a novel locus for intellectual disability. Hum. Genet. 135, 773–778 (2016).
Segawa, K. et al. A sublethal ATP11A mutation associated with neurological deterioration causes aberrant phosphatidylcholine flipping in plasma membranes. J. Clin. Investig. https://doi.org/10.1172/JCI148005 (2021).
Walhovd, K. B., Watts, R., Amlien, I. & Woodward, L. J. Neural tract development of infants born to methadone-maintained mothers. Pediatr. Neurol. 47, 1–6 (2012).
Perron, J. C., Rodrigues, A. A., Surubholta, N. & Dodd, J. Chemotropic signaling by BMP7 requires selective interaction at a key residue in ActRIIA. Biol. Open https://doi.org/10.1242/bio.042283 (2019).
Al Turki, S. et al. Rare variants in NR2F2 cause congenital heart defects in humans. Am. J. Hum. Genet. 98, 592 (2016).
Law, R. et al. Biallelic truncating mutations in FMN2, encoding the actin-regulatory protein Formin 2, cause nonsyndromic autosomal-recessive intellectual disability. Am. J. Hum. Genet. 95, 721–728 (2014).
Gellens, M. E. et al. Primary structure and functional expression of the human cardiac tetrodotoxin-insensitive voltage-dependent sodium channel. Proc. Natl. Acad. Sci. U. S. A. 89, 554–558 (1992).
Mujtaba, S., Romero, J. & Taub, C. C. Methadone, QTc prolongation and torsades de pointes: Current concepts, management and a hidden twist in the tale?. J. Cardiovasc. Dis. Res. 4, 229–235 (2013).
Parikh, R., Hussain, T., Holder, G., Bhoyar, A. & Ewer, A. K. Maternal methadone therapy increases QTc interval in newborn infants. Arch. Dis. Child. Fetal Neonatal Ed. 96, F141–F143 (2011).
Luo, X., Kranzler, H. R., Zhao, H. & Gelernter, J. Haplotypes at the OPRM1 locus are associated with susceptibility to substance dependence in European-Americans. Am. J. Med. Genet. B Neuropsychiatr. Genet. 120B, 97–108 (2003).
Leventelis, C. et al. Buprenorphine and methadone as opioid maintenance treatments for heroin-addicted patients induce oxidative stress in blood. Oxid. Med. Cell. Longev. 2019, 9417048 (2019).
Tsai, M.-C. & Huang, T.-L. Brain-derived neurotrophic factor (BDNF) and oxidative stress in heroin-dependent male patients undergoing methadone maintenance treatment. Psychiatry Res. 249, 46–50 (2017).
Obst, S. et al. Perinatal hyperoxia and developmental consequences on the lung-brain axis. Oxid. Med. Cell. Longev. 2022, 5784146 (2022).
Vestal-Laborde, A. A., Eschenroeder, A. C., Bigbee, J. W., Robinson, S. E. & Sato-Bigbee, C. The opioid system and brain development: Effects of methadone on the oligodendrocyte lineage and the early stages of myelination. Dev. Neurosci. 36, 409–421 (2014).
Domené, H. M. & Fierro-Carrión, G. Genetic disorders of GH action pathway. Growth Horm. IGF Res. 38, 19–23 (2018).
Holt, R. I. G. Fetal programming of the growth hormone–insulin-like growth factor axis. Trends Endocrinol. Metab. 13, 392–397 (2002).
Sasaki, A., Murphy, K. E., Briollais, L., McGowan, P. O. & Matthews, S. G. DNA methylation profiles in the blood of newborn term infants born to mothers with obesity. PLoS One 17, e0267946 (2022).
Lorente-Pozo, S. et al. DNA methylation analysis to unravel altered genetic pathways underlying early onset and late onset neonatal sepsis. A pilot study. Front. Immunol. 12, 622599 (2021).
Fong, G. et al. DNA methylation profile in human cord blood mononuclear leukocytes from term neonates: Effects of histological chorioamnionitis. Front. Pediatr. 8, 437 (2020).
Gayen Nee ‘Betal, S. et al. Histological chorioamnionitis induces differential gene expression in human cord blood mononuclear leukocytes from term neonates. Sci. Rep. 9, 5862 (2019).
Acknowledgements
The authors acknowledge the support of Cancer Genomics Facility, Sidney Kimmel Cancer Center, and the Hemant Desai Grant Committee, Thomas Jefferson University, Philadelphia.
Funding
This work was funded by Hemant Desai Fellow Research Grant, Thomas Jefferson University, (102-24000-7200048).
Author information
Authors and Affiliations
Contributions
O.A: Sample collection, processing, data analysis, manuscript write up and editing. SGB: Sample collection and processing, data interpretation, manuscript write up and editing. PU: Data collection and manuscript writing. RH: Data collection and manuscript writing. KB: Data collection and manuscript writing. HBA: Concept and design, data interpretation manuscript writing. KS: Concept and design, data interpretation, manuscript writing. JSC: Concept and design, data interpretation, placental tissue analysis, manuscript writing. SA: Concept and design, data collection, sample processing, data analysis and interpretation, and manuscript writing. RCB: concept and design, data interpretation, manuscript writing. Z.A: Concept and design, data analysis and interpretation, manuscript writing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Adegboyega, O., Gayen nee’ Betal, S., Urday, P. et al. DNA methylation patterns in umbilical cord blood from infants of methadone maintained opioid dependent mothers. Sci Rep 14, 17298 (2024). https://doi.org/10.1038/s41598-024-66899-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-66899-w
- Springer Nature Limited