Abstract
Physical mapping evidences the chromosome organization and structure. Despite the data about plant cytogenomics, physical mapping has been conducted from single-copy and/or low-copy genes for few species. Carica papaya cytogenomics has been accomplished from BAC-FISH and repeatome sequences. We aimed to map the serk 2, svp-like and mdar 4 sequences in C. papaya. The sequences were amplified and the amplicons sequenced, showing similarity in relation to serk 2, svp-like and mdar 4 genes. Carica papaya diploidy was confirmed and the mitotic chromosomes characterized. The chromosome 1 exhibited the secondary constriction pericentromeric to the centromere of the long arm. So, we concluded that it is the sex chromosomes. serk 2 was mapped in the long arm interstitial portion of the sex chromosomes, and the interphase nuclei showed two fluorescence signals. Considering these results and the sequencing data from the C. papaya sex chromosomes, svp-like and mdar 4 genes were mapped in the interstitial region of the sex chromosome long arm. Both sequences showed only one fluorescence signal in the interphase nuclei. The procedure adopted here can be reproduced for other single-copy and/or low-copy genes, allowing the construction of cytogenetic maps. In addition, we revisited the cytogenomics data about C. papaya sex chromosomes, presenting a revised point of view about the structure and evolution to these chromosomes.
Similar content being viewed by others
Introduction
Carica papaya L. (Caricaceae) originated in Mesoamerica. This species is an important commercial fruit cultivated in tropical and subtropical regions of the world1. Seminal propagation is the main system used for C. papaya cultivation, resulting in individuals of three biological sexes typically following a Mendelian inheritance2. Carica papaya possesses a relatively small nuclear genome size showing ~ 1C = 0.32 pg, equivalent to ~ 318 Mbp3. This species is diploid with karyotype exhibiting 2n = 2x = 18 chromosomes, which have been classified as metacentric and submetacentric3,4.
Sex chromosome evolution and sexual differentiation mechanisms have been investigated from different plant species, and C. papaya stands out as an important model species5. Sex differentiation in C. papaya was initially based on one gene with at least three alleles (M1, M2 and m). Considering this proposal, male, hermaphrodite and female plants differentiated from the genotypes M1 m, M2 m and m m, respectively6,7. The genotypes M1 M1, M2 M2 and M2 M1 are lethal, promoting the zygotic embryo death6,7. Storey revised his hypothesis, suggesting that the sex is not differentiated by the expression of one gene with three alleles, but by linked genes located in a region of the sex chromosomes where recombination is suppressed8. Corroborating to this Storey’s hypothesis, further evidence showed that the recombination is severely suppressed in the region close to the sexual differentiation locus9, and at least seven genes were identified in the sexual specific portion10. Furthermore, structural chromosomal rearrangements (inversions, deletions and translocations), which may be the outcomes of the recombination suppression, were detected in the sex differentiation region10.
Sex differentiation region in C. papaya genome was genetically mapped to linkage group 1, which is related to the chromosome 111,12. C. papaya biological sex is expressed by a XY chromosome sex differentiation system, from which females are homogametic (XX), and males (XY) and hermaphrodites (XYh) are heterogametic. Y and Yh chromosomes have a specific sexual differentiation region that shows few genetic differences13. Male-specific region (MSY) and hermaphrodite-specific region (HSY) are about 9.8 Mbp, while X-specific region (XX) has 6.0 Mbp14. MSY and HSY chromosome portions have a larger genome size in relation to the corresponding region on the X chromosome, mainly due to the DNA sequence duplications and insertion of mobile elements (retrotransposons) in this region13. HSY possesses more repetitive sequences than X. In addition, 121 pseudoautosomal genes occur between HSY and X chromosomes, 56 specific HSY genes, and 74 specific X genes14.
In association to cytogenomics, genomic sequencing data makes possible the physical mapping in mitotic and meiotic chromosomes of different DNA sequences, including single-copy and/or low-copy genes. Fluorescent in situ hybridization (FISH) is a cytogenomics technique that maps DNA sequences on chromosomes. FISH contributes to the construction of physical maps, and, consequently, the integration of these maps with genetic maps15,16. FISH in C. papaya 'Solo' and 'Maradol' mitotic chromosomes from 18S and 5S rDNA sequences evidenced the 18S rDNA in the pericentromeric portion of one chromosome pair, while the 5S rDNA was mapped in the pericentromeric portion of three chromosome pairs17. In contrast, pachytene chromosomes of C. papaya 'SunUp' showed 5S rDNA on chromosomes 3, 5, 8, 9 and Y, and the 45S rDNA was mapped in the pericentromeric region of the chromosome 4 short arm15. BAC-FISH mapped the BAC clones associated with the 12 linkage groups (LG) on the C. papaya pachytene chromosomes: BAC 96C17 (LG 1), BAC 39C20 (LG 9) and BAC 39P03 (LG 11), BAC 23B18 (LG 6 ), BAC 15O14 (LG 2), BAC 57E17 (LG 5), BAC 07H21 (LG 3), BAC 12M21 (LG 7) and BAC 01P02 (LG 12), BAC 43N18 (LG 4), BAC 78D03 (LG 8) and BAC 99D21 (LG 10), respectively on chromosomes 1 (XY), 2, 3, 4, 5, 6, 7, 8 and 9, integrating the LG to the individual chromosomes15.
In plants, physical mapping has contributed to the characterization of the sex chromosomes. The mapping of a repetitive DNA sequence, named HSR1, in Humulus lupulus L. showed that this sequence is located in the subtelomeric region of the X and Y chromosome long arm. However, the HSR1 was also mapped in the pericentromeric portion of the chromosome X18. In Cannabis sativa L., the repetitive sequence named CS-1 was mapped in the subtelomeric region of the Y chromosome short arm, while both arms of the X chromosome showed this sequence in the subtelomeric region19. The mapping of repetitive sequences in Hippophae rhamnoides L. revealed the HRTR 12 repetitive sequence only in the Y chromosome20. Repetitive DNA sequences, named RAYS, were only mapped in the Y chromosome of Rumex acetosa L.21. Therefore, the sex chromosome characterization in plants has been conducted mainly from the repeatome sequences. In addition to physical mapping, FISH has been accomplished to assists in the early identification of sporophyte sex. For example, a polymorphic molecular marker named NAPF-2 showed fluorescent signals in leaf nuclei of hermaphrodite plants, however, no signal was detected in leaf nuclei of female plants of C. papaya ‘Golden’ and ‘Rubi’22.
In addition to these sequences (mainly from the repeatoma), the physical mapping of single-copy and/or low-copy genes is needed to expand the knowledge about the C. papaya genome, contributing to knowing and understanding its organization and evolution. Moreover, these genes also have potential as cytomolecular markers. In this context, some sequences that occur in the sexual differentiation region were explored here, including: somatic embryogenesis receptor kinase gene (serk 2), short vegetative phase gene (svp-like) and monodehydroascorbate reductase gene (mdar 4)12,23.
The serk 2 gene, which was sequenced in the X and Yh chromosomes14, encodes a protein that belongs to the plasma membrane receptor kinase family. SERK protein has leucine-rich repeats, acting on signal transduction23 and male sporogenesis24. The svp-like gene encodes a transcription factor that regulates the transition from the vegetative to the reproductive phase, activating the classes B and C genes of the floral morphogenesis25,26. Gene expression and sequencing approaches showed that the svp-like gene present on the Y chromosome encodes a wild-type protein with all domains. Due to a Copia-like retrotransposon insertion, a mutant allele of this gene occurs on the Yh chromosome, encoding a protein that has only the K-box domain12,13,27,28. In addition, svp-like gene was also identified on the chromosome 414. The mdar 4 gene, which was also sequenced in X and Yh chromosomes14, encodes an enzyme that has antioxidant activity, eliminating reactive oxygen species and, consequently, increasing tolerance against oxidative stress in some plant species29,30. Genetic analyzes have shown that the mdar 4 gene has a wild-type allele on the X chromosome, while the allele of the Y and Yh chromosomes contains a LTR-retroelement sequence30. Y and Yh mutant alleles encode a shorter truncated MDAR 4 enzyme in flower tissues28.
Considering the previous genomic and cytogenomic data, we hypothesize that C. papaya sex chromosomes differ in several DNA sequences31, including svp-like and mdar 4 genes. Based on this, we aimed to map the serk 2, svp-like and mdar 4 sequences in C. papaya mitotic chromosomes. So, we start the mapping of single-copy and/or low-copy genes, differing from the previous C. papaya cytogenomics that were accomplished from rDNA genes and BAC sequences.
Results
The immersion of seeds in GA3 solution provided a high germination rate (95%) after ~ 5 days at 30 °C. Break seed dormancy was essential to increase the germination rate in a short period, providing a large amount of root meristems to obtain mitotic cells. Prometaphases and metaphases with morphologically preserved chromosomes exhibiting well-defined primary and secondary constrictions were obtained from the cytogenetics procedure. Therefore, we considered adequate the root meristem treatment with amiprophos-methyl/dimethyl sulfoxide, the enzymatic maceration, and the slide preparation accomplishing macerated root dissociation and air drying. The prometaphases and metaphases showed 2n = 2x = 18 chromosomes, confirming the C. papaya diploidy. The mean total chromosome length ranged from 4.32 ± 1.53 µm (chromosome 1) to 2.70 ± 0.42 µm (chromosome 9). Carica papaya karyogram possesses four metacentric (1, 5, 7 and 8) and five submetacentric chromosomes (2, 3, 4, 6 and 9, SI Table 1). Secondary constriction was identified in the pericentromeric region of the chromosome 1 (Fig. 1).
Physical mapping of the serk 2, mdar 4 and svp-like sequences in mitotic chromosomes of C. papaya. Detail for the markings of the serk 2, mdar 4 and svp-like in the interstitial region of the chromosome 1 long arm. The karyograms show 2n = 2x = 18 chromosomes, aligned by the centromere. The secondary constriction present in the pericentromeric region of chromosome 1 is observed. Bars: 10 μm.
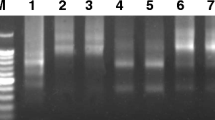
Sequences with ~ 1,100, ~ 250 and ~ 700 bp were amplified from C. papaya genomic DNA and serk 2, mdar 4 and svp-like primers, respectively (SI Fig. 1). The similarity of the amplified serk 2 sequence in relation to C. papaya ‘Sunset’ and ‘SunUp’ genomes14 was 85% for sense strand and 82% for antisense strand in relation to sequenced chromosome 1, and 83% for sense strand and 81% for antisense strand in relation to chromosome Yh. For mdar 4 sequence, the similarity was 95% for sense strand and 94% for antisense strand in relation to the chromosome 1, and 90% for sense strand and 89% for antisense strand in relation to the chromosome Yh. The similarity of the svp-like sequence was 100% for sense strand and 99% for antisense strand in relation to the chromosome 4, and 86% for sense strand and 82% for antisense strand in relation to the chromosome Yh.
We defined the FISH procedure based on different tests and their respective results, considering the visualization of fluorescence signals in nuclei and mitotic chromosomes. We used 4X SSC in the post-hybridization wash solution (stringency). Thus, the FISH conditions were established for the serk 2, svp-like and mdar 4 probes, which showed different sizes in bp. The hybridizations were performed in prometaphases and metaphases to map these sequences, and also in interphase nuclei to confirm the copy number of each sequence. So, clear signals were generated after hybridization of the probes in nuclei and chromosomes.
The serk 2 sequence was mapped in prometaphases/metaphases on the interstitial region of the long arm of chromosome 1 pair, which was identified from the secondary constriction in pericentromeric region. This chromosome has been appointed as the sex chromosome of C. papaya. In addition, two fluorescence signals were detected in interphase nuclei, confirming that there is one copy of the serk 2 in the C. papaya genome. As well as serk 2, svp-like and mdar 4 sequences were mapped in prometaphases/metaphases in the interstitial region of the long arm of chromosome 1. However, only one chromosome 1 exhibited these sequences (Figs. 1 and 2). Corroborating this, we found one svp-like and mdar 4 fluorescence in interphase nuclei. Thus, our results suggest differences between the chromosomes of the homologous pair 1, also designated as X and Y/Yh.
Based on the karyograms (Fig. 1), we structured an ideogram of the chromosomes X and Y/Yh, representing the loci marked by the serk 2, mdar 4 and svp-like, as well as the centromere and the secondary constriction (Fig. 3). The results obtained in this study increase the cytogenomic data of C. papaya through the physical mapping of the serk 2, mdar 4 and svp-like. In addition, the ideogram includes the cytogenomic data of the physical location of heterochromatic portions32, the 5S rDNA gene15 and the distribution of retroelements27.
Ideograms of the X and Yh/Y chromosomes of C. papaya. Chromosomes 1 exhibiting the fluorescence signal for serk 2, mdar 4 and svp-like sequences. The orange bar represents the position of the centromere. The light blue circle shows the position of the secondary constriction. The pink and purple circles on the long arm of the X chromosome show the loci of the serk 2 and mdar 4 sequences, respectively. The pink, purple and red circles on the long arm of the Yh/Y chromosome shows the loci of the serk 2, mdar 4 and svp-like genes. The yellow circles represent the knobs (heterochromatic portions)39. The dark blue circle in the pericentromeric region of the long arm of the Y chromosome represents the 5S rDNA15. The green circles show the distribution of retroelements along the sex chromosomes27.
Discussion
We mapped the serk 2, mdar 4 and svp-like, providing the first physical mapping of these sequences in C. papaya mitotic chromosomes. Accomplishing the same FISH procedure, we mapped amplicons from 251 to 1166 bp, resulting in a physical mapping in mitotic chromosomes from sequences of single-copy or low-copy in C. papaya. Our procedure can be reproduced allowing the physical mapping of genes with single-copy or low-copy. In addition, our results represent the start point for further physical mapping, which are relevant to increase knowledge about the C. papaya genome. We integrated the physical map with the in silico data, contributing to knowing and understanding of the sexual chromosomes X and Y/Yh of the C. papaya. In addition, the svp-like can be further used as cytomolecular markers to previously screen the sex of C. papaya plants.
C. papaya is a model species for studies about sex chromosome evolution and sex differentiation in plants, mainly due to incipient evolution of its sex chromosomes X and Y/Yh, and the recombination suppression in the non-homologue portion between them1,5. In addition to the svp-like and mdar 4 sequences, a relatively high density of retroelements was identified along the C. papaya sex chromosomes27. Furthermore, at least four knobs (heterochromatic portions) and highly methylated sequences were identified only in the sexual region of the Y/Yh chromosome and not in the non-homologous portion of the X chromosome32. These cytogenomic characteristics may influence gene expression and sequence divergence.
serk 2 physical mapping revealed its locus in the interstitial region of the chromosome 1 long arm. The sequencing of the X and Y/Yh sex chromosome dedifferentiation portions showed that the serk 2 gene is present in this region12,14. The presence of two signals in the nuclei and in the metaphases confirms that the serk 2 is a single-copy gene, and that it possibly occurs near or in the pseudoautosomal portion of the X and Y/Yh chromosomes. Although the sex determination region has specific genes or sequences of each sex chromosome (74 genes in X, and 56 genes in Yh), at least 121 genes have been predicted in the sex region of the X chromosome and in its Yh counterpart13,14.
mdar 4 sequence, which was amplified from C. papaya genomic DNA, locus was also mapped in the chromosome 1 interstitial region, and only one hybridization signal of this sequence was detected in the nuclei and chromosomes. Genomic analyses showed that the divergence between the mdar 4 gene in sex chromosomes is due to the insertion of retroelements inside of this gene in Y (male) and Yh (hermaphrodite) chromosomes, which does not occur in X chromosome27. Since the mdar 4 gene plays a role in eliminating reactive oxygen species, maintaining cell viability, the absence of this gene on the X chromosome can be lethal30. The alignment of a region of the X and Yh C. papaya chromosomes revealed deletions on the Yh. For example, the asymmetric leaves 2 gene appears to have been lost, since it occurs only on the X chromosome10. Gene loss has been reported in Silene latifolia Poir. Y chromosome, which lossed ~ 14.5 of genes33. Thus, further studies in C. papaya, including cytogenomics, need to be conducted to show possible gene loss between the X and Y/Yh chromosomes.
svp-like was mapped in the interstitial region of the 1 chromosome long arm. The transcriptome indicated that svp-like gene occurs only on the sex differentiation region of the Y (male) and Yh (hermaphrodite) chromosome27. svp-like gene was also identified by sequencing in the chromosome 414. The sequencing showed that our svp-like sequence possesses similarity in relation to svp-like gene of the Yh chromosome. Therefore, our data suggest that the svp-like sequence was mapped in the Y/Yh chromosome.
The mapping of the svp-like and mdar 4 sequences on chromosome 1, which shows the secondary constriction and the higher total length. So, this result corroborates with sex differentiation region in C. papaya, which is in a pericentromeric region that expands to the long arm of this chromosome33. The expansion of the suppressed recombination region occurs from the selection of sexual antagonistic genes and/or chromosomal rearrangements33,34,35. As a consequence of the recombination suppression in the sex differentiation region, the Y/Yh chromosome undergoes genetic degeneration, presenting low expression or loss of genes. Suppressed recombination in sex differentiation region can promote divergence between the sequences present in this chromosomal portion and gradually change its copy number.
The identification of cytogenetic differences and the characterization of sex chromosomes in plants have been conducted mainly through physical mapping of repetitive sequences. In H. lupulus, the satellite DNA HSR1 was mapped in the subtelomeric and pericentromeric region of the X chromosome, while the Y chromosome presented this sequence only in the subtelomeric region18. The mapping of the repetitive sequence CS-1 differentiated the sex chromosomes of C. sativa, due to this sequence occurs only in the subtelomeric region of the Y chromosome short arm and in the subtelomeric region of both arms of the X chromosome18. In H. rhamnoides and R. acetosa, the satellite DNA sequences HRTR 12 and RAYS, respectively, were mapped only on the Y chromosome20,21.
In addition to the sex chromosomes characterization, molecular cytogenetics have been accomplished to determine the plant sex before the reproductive period. Based on our results and previous sequencing data, the svp-like sequence can be explored as cytomolecular markers to early determine the sex of C. papaya plants. In C. papaya, molecular markers of the Sequence Characterized Amplified Region type (SCAR) were used to differentiate hermaphrodite individuals from females, through the FISH application in nuclei isolated from the leaves. Using the molecular marker NAPF-2, only the nuclei isolated from the hermaphrodite plants showed a strong fluorescence signal, which was not observed in the leaf nuclei of female plants22. Thus, FISH is also useful to early determine and screen the sex, reducing the growing costs of the sex of noncommercial interest.
For the first time, single-copy sequences were mapped in C. papaya sex chromosomes. We mapped serk 2, svp-like and mdar 4 sequences, which are single-copy or low-copy sequences, in mitotic chromosomes of C. papaya. Therefore, our data represents the start point to map other specific sex chromosome genes, increasing cytogenomics analyzes and the understanding of the evolution and structure of the sex chromosomes in C. papaya.
Material and methods
Commercial C. papaya ‘Hawaii’ (Isla®), a gynodioecious line, seeds were bought in accordance to relevant guidelines and legislation. We declare that all methods were performed in accordance with the relevant guidelines.
Data availability
The datasets analysed for serk 2, svp-like and mdar 4 sequences during the current study are available in: https://www.ncbi.nlm.nih.gov/gene/?term=LOC110813265 for serk 2, https://www.ncbi.nlm.nih.gov/nuccore/XM_022047757.1 for svp-like, and https://www.ncbi.nlm.nih.gov/gene/?term=LOC110807062 for mdar 4, as well as https://www.nature.com/articles/s41588-022-01068-1, and https://ngdc.cncb.ac.cn/gwh/ GWHBFSC00000000 for C. papaya ‘SunUp’ and GWHBFSD00000000 for C. papaya ‘Sunset’14 for the three genes. The chromosome morphometry data generated and analysed during this study are included in this published article and its supplementary information files.
Mitotic chromosomes
Commercial seeds of C. papaya ‘Hawaii’ (Isla®) were immersed in 200 ppm gibberellic acid (GA3) for 24 h to break seed dormancy36. Subsequently, the seeds were placed in Petri dishes with filter paper moistened with dH2O and kept at 30 °C until germination. The 0.5–1.0 cm long roots were treated with 3 µM amiprophos-methyl (Sigma®) and 0.3% dimethyl sulfoxide (Sigma®) for 4 h at 30 °C. The roots were washed in dH2O and fixed in methanol:acetic acid (3:1), with three changes of 10 min each, and kept at − 20 °C37,38. Root meristems were excised, washed in dH2O three times, and incubated at 36 °C for 2 h in an enzyme solution (4% cellulase Sigma®, 0.4% hemicellulase Sigma®, 1% macerozyme R10 Yakult®, 100% pectinase Sigma®) diluted in dH2O at 1:14 (enzymatic solution: dH2O). After enzymatic maceration, root meristems were washed in dH2O, fixed in methanol:acetic acid (3:1) and kept at − 20 °C. The slides were prepared using cell dissociation and air-drying techniques3.
FISH
Carica papaya total genomic DNA was extracted from young leaves using GenElute™ Plant Genomic DNA Miniprep Kit (Sigma®), following the manufacturer's instructions. DNA concentration and purity were determined by a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific®), and its integrity was evaluated by electrophoresis in a 1.5% agarose gel. The primers of the serk 2 and mdar 4 were designed based on the sequences deposited at the National Center for Biotechnology Information (NCBI): serk 2—LOC110813265 and mdar 4—LOC110807062. For the svp-like, the primers were obtained based on the bibliography34. So, we designed the primers: F 5′-CTCTCACTGCACGCCTAAC-3′ and R 5′-TCGCCTTCAAATCCTGAAACT-3' for serk 2 providing a 1,166 bp amplification product; F 5′-ACTTGTTGCCTCAGTTTCTCATTCTCTTC-3′ and R 5′-GAGATCAGTGATCTTCAAAGGAAGGTC-3′ for svp-like resulting a 251 bp amplification product; and F 5′-TATTCCGACCCCAGTCTCCA-3' and R 5'-TCCTACCGCGCCAAACAAAT-3′ for mdar 4 providing a 731 bp amplification product. The primers were validated in silico using the OligoAnalyzer™ tool (https://www.idtdna.com/pages/tools/oligoanalyzer), and the formation of homodimers, heterodimers and hairpin structures were verified. Specificity analysis was performed using the Basic Local Alignment Search Tool (BLAST, https://blast.ncbi.nlm.nih.gov/Blast.cgi).
PCR reactions for amplification the gene sequences consisted of 0.5 μM of each primer, 60 ng genomic DNA, 200 μM of each dNTP (Promega®), 1.3 mM MgCl2 (Promega®), 1X Colorless GoTaq® Flexi Buffer (Promega®) and 1 U of the enzyme GoTaq® Flexi DNA Polymerase (Promega®). All amplifications were performed in a PTC-200 Peltier Thermal Cycler (MJ Research, Inc) under the following conditions: 4 min at 95 °C, followed by 30 cycles of 1 min at 95 °C, 1 min at 54 °C, 50 °C and 50 °C, respectively, for the serk 2, svp-like and mdar 4 primers, 1 min at 72 °C, and 5 min at 72 °C for a final extension. PCR products were analyzed on 1.5% agarose gel (SI Fig. 1).
PCR reactions for probe labeling were accomplished from amplified gene fragments. The reactions consisted of 0.5 μM of each primer, 200 ng of previously amplified sequences, 200 μM of each dATP, dCTP and dGTP (Promega®), 100 μM of dTTP (Promega®), 20 μM Tetramethylrhodamine-5-dUTP (Roche®), 1.3 mM MgCl2, 1X Colorless GoTaq® Flexi Buffer (Promega®) and 1 U of the enzyme GoTaq® Flexi DNA Polymerase (Promega®)38. The PCR conditions were the same used for the gene sequences amplification.
Slides with prometaphases and metaphases exhibiting chromosomes with well-defined centromeres were selected from phase contrast microscope Olympus BX41. The selected slides were treated in 1X phosphate-buffered saline (PBS) for 5 min, fixed in 4% formalin solution for 10 min, and washed with 1X PBS for 5 min. Subsequently, the slides were dehydrated in an ice-cold alcoholic series (70%, 85% and 100%) for 3 min each. The hybridization mix consisted of 200 ng of each probe, 50% formamide and 2X saline sodium citrate (SSC). Chromosomes were denatured in a water bath at 70 °C for 3 min in 70% formamide/2X SSC. After denaturation, the slides were dehydrated in an ice-cold alcoholic series (70%, 85% and 100%) for 3 min each. The hybridization mix was denatured in a PTC-200 Peltier Thermal Cycler (MJ Research, Inc) at 85 °C for 5 min and immediately placed on ice for at least 5 min. The hybridization mix was placed on the slides, covered with HybriSlip™ plastic coverslips (Sigma®) and sealed with Rubber Cement glue (Elmer’s®). Hybridization was carried out in the ThermoBrite™ equipment (StatSpin®) at 37 °C for 24 h. Subsequently, the slides were submitted to a stringency wash performed at 40 °C in 4X SSC solution for 5 min. Slides were counterstained with 10% glycerol/PBS + 4′,6-diamidino-2-phenylindole (DAPI), covered with a glass coverslip and sealed with nail polish39,40,41,42. Nuclei, prometaphases and metaphases were captured with a 12-bit CCD digital camera (Olympus®) attached to an Olympus BX-60 photomicroscope equipped with epifluorescence and 100 × PlanApo immersion objective (NA = 1.4). Captured images were processed using Image-Pro Plus 6.1 software (Media Cybernetics, Inc).
Sequencing of the amplification products from serk 2, svp-like and mdar 4 primers
PCR reactions were performed with a total volume of 50 μL and consisted of 0.5 μM of each primer, 200 ng of previously amplified sequences, 200 μM of each dNTP (Promega®), 1.3 mM of MgCl2, 1X of Colorless GoTaq® Flexi Buffer (Promega®) and 1 U of the enzyme GoTaq® Flexi DNA Polymerase (Promega®). PCR conditions were the same used for amplification of the sequences from serk 2, svp-like and mdar 4 primers. The purification of the amplified sequences was performed with the Wizard® SV Gel and PCR Clean-Up System (Promega®) kit, following the manufacturer’s instructions. Concentration and purity of the PCR products were determined by a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific®), and their integrity was evaluated by electrophoresis in a 1.5% agarose gel. Purified PCR products were sent for sequencing by the Sanger method at ACTGene Análises Moleculares.
From sequenced serk 2, svp-like and mdar 4, we verified the similarity of these sequences in relation to the C. papaya ‘SunUp’ (JAIUCH000000000, https://www.ncbi.nlm.nih.gov/nuccore/JAIUCH010000010.1) and C. papaya ‘Sunset’ (JAIUCG000000000, https://www.ncbi.nlm.nih.gov/nuccore/JAIUCG010000010.1) genomes14. We evaluated the similarity of our amplified serk 2, svp-like and mdar 4 sequences in relation to each C. papaya chromosome: chromosome 1 that contains the X and Yh sexual differentiation regions, chromosomes 2–9, and chromosome Yh (SI Genomic Data).
Data availability
All data generated and analysed during this study are included in this published article and its supplementary information files.
References
Lee, C. Y. et al. The development of functional mapping by three sex-related loci on the third whorl of different sex types of Carica papaya L.. PLoS ONE 13, e0194605. https://doi.org/10.1371/journal.pone.0194605 (2018).
Costa, A. F. S., Abreu, E. F. M., Schmildt, E. R., Costa, A. N. & Schmildt, O. Advances observed in papaya tree propagation. Rev. Bras. Frutic. https://doi.org/10.1590/0100-29452019036 (2019).
Araújo, F. S., Carvalho, C. R. & Clarindo, W. R. Genome size, base composition and karyotype of Carica papaya L.. Nucleus 53, 25–31. https://doi.org/10.1007/s13237-010-0007-8 (2010).
Damasceno Junior, P. C., da Costa, F. R., Pereira, T. N. S., Neto, M. F. & Pereira, M. G. Karyotype determination in three Caricaceae species emphasizing the cultivated form (C. papaya L.). Caryologia 62, 10–15. https://doi.org/10.1080/00087114.2004.10589660 (2009).
Aryal, R. & Ming, R. Sex determination in flowering plants: papaya as a model system. Plant Sci. 217, 56–62. https://doi.org/10.1016/j.plantsci.2013.10.018 (2014).
Hofmeyr, J. D. J. Genetical studies of Carica papaya L.. S. Afr. J. Sci. 35, 300–304 (1938).
Storey, W. B. Segregations of sex types in Solo papaya and their application to the selections of seed. J. Am. Soc. Hortic. Sci. 35, 83–85 (1938).
Storey, W. B. Genetics of the papaya. J. Hered. 44, 70–78. https://doi.org/10.1093/oxfordjournals.jhered.a106358 (1953).
Ma, H. et al. High-density linkage mapping revealed suppression of recombination at the sex determination locus in papaya. Genetics 166, 419–436. https://doi.org/10.1534/genetics.166.1.419 (2004).
Yu, Q. et al. Low X/Y divergence in four pairs of papaya sex-linked genes. Plant J. 53, 124–132. https://doi.org/10.1111/j.1365-313X.2007.03329.x (2008).
Ming, R., Yu, Q. & Moore, P. H. Sex determination in papaya. Semin. Cell Dev. Biol. 18, 401–408. https://doi.org/10.1016/j.semcdb.2006.11.013 (2007).
VanBuren, R. et al. Origin and domestication of papaya Yh chromosome. Genome Res. 25, 524–533. https://doi.org/10.1101/gr.183905.114 (2015).
Wang, J. et al. Sequencing papaya X and Yh chromosomes reveals molecular basis of incipient sex chromosome evolution. Proc. Natl. Acad. Sci. U.S.A. 109, 13710–13715. https://doi.org/10.1073/pnas.1207833109 (2012).
Yue, J. et al. SunUp and Sunset genomes revealed impact of particle bombardment mediated transformation and domestication history in papaya. Nat. Genet. 54, 715–724. https://doi.org/10.1038/s41588-022-01068-1 (2022).
Zhang, W., Wai, C. M., Ming, R., Yu, Q. & Jiang, J. Integration of genetic and cytological maps and development of a pachytene chromosome-based karyotype in papaya. Trop. Plant Biol. 3, 166–170. https://doi.org/10.1007/s12042-010-9053-2 (2010).
Yu, Q. Physical map of papaya genome. In Genetics and Genomics of Papaya (eds Ming, R. & Moore, P. H.) 169–183 (Springer, 2014).
Costa, F. R., Pereira, T. N. S., Hodnett, G. L., Pereira, M. G. & Stelly, D. M. Fluorescent in situ hybridization of 18S and 5S rDNA in papaya (Carica papaya L.) and wild relatives. Caryologia 61, 411–416. https://doi.org/10.1080/00087114.2008.10589655 (2008).
Divashuk, M. G., Alexandrov, O. S., Kroupin, P. Y. & Karlov, G. I. Molecular cytogenetic mapping of Humulus lupulus sex chromosomes. Cytogenet. Genome Res. 134, 213–219. https://doi.org/10.1159/000328831 (2011).
Divashuk, M. G., Alexandrov, O. S., Razumova, O. V., Kirov, I. V. & Karlov, G. I. Molecular cytogenetic characterization of the dioecious Cannabis sativa with an XY chromosome sex determination system. PLoS One. 9, e85118. https://doi.org/10.1371/journal.pone.0085118 (2014).
Puterova, J. et al. Satellite DNA and transposable elements in seabuckthorn (Hippophae rhamnoides), a dioecious plant with small Y and large X chromosomes. Genome Biol. Evol. 9, 197–212. https://doi.org/10.1093/gbe/evw303 (2017).
Jesionek, W. et al. Fundamentally different repetitive element composition of sex chromosomes in Rumex acetosa. Ann. Bot. 127, 33–47. https://doi.org/10.1093/aob/mcaa160 (2021).
Abreu, I. S., Carvalho, C. R. & Soares, F. A. Early sex discrimination in Carica papaya by nuclei FISH. Euphytica 206, 667–676. https://doi.org/10.1007/s10681-015-1485-1 (2015).
Ming, R. et al. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 452, 991–996. https://doi.org/10.1038/nature06856 (2008).
Albrecht, C., Russinova, E., Hecht, V., Baaijens, E. & de Vries, S. The Arabidopsis thaliana somatic embryogenesis receptor-like kinases1 and 2 control male sporogenesis. Plant Cell 17, 3337–3349. https://doi.org/10.1105/tpc.105.036814 (2005).
Gregis, V., Sessa, A., Colombo, L. & Kater, M. M. Agl 24, short vegetative phase, and apetala 1 redundantly control agamous during early stages of flower development in Arabidopsis. Plant Cell 18, 1373–1382. https://doi.org/10.1105/tpc.106.041798 (2006).
Gregis, V. et al. Identification of pathways directly regulated by short vegetative phase during vegetative and reproductive development in Arabidopsis. Genome Biol. 14, 1–26. https://doi.org/10.1186/gb-2013-14-6-r56 (2013).
Urasaki, N. et al. Digital transcriptome analysis of putative sex-determination genes in papaya (Carica papaya). PLoS One. 7, e40904. https://doi.org/10.1371/journal.pone.0040904 (2012).
Zerpa-Catanho, D., Clough, S. J. & Ming, R. Characterization and analysis of the promoter region of monodehydroascorbate reductase 4 (CpMDAR4) in papaya. Plant Reprod. 35, 233–264. https://doi.org/10.1007/s00497-022-00447-2 (2022).
Ueno, H. et al. Genome sequence comparison reveals a candidate gene involved in male–hermaphrodite differentiation in papaya (Carica papaya) trees. Mol. Genet. Genom. 290, 661–670. https://doi.org/10.1007/s00438-014-0955-9 (2015).
Eltayeb, A. E. et al. Overexpression of monodehydroascorbate reductase in transgenic tobacco confers enhanced tolerance to ozone, salt and polyethylene glycol stresses. Planta 225, 1255–1264. https://doi.org/10.1007/s00425-006-0417-7 (2007).
Ávila-Hernández, J. G., del Rosario Cárdenas-Aquino, M., Camas-Reyes, A. & Martínez-Antonio, A. Sex determination in papaya: Current status and perspectives. Pant Sci. 335, 1–10. https://doi.org/10.1016/j.plantsci.2023.111814 (2023).
Zhang, W., Wang, X., Yu, Q., Ming, R. & Jiang, J. DNA methylation and heterochromatinization in the male-specific region of the primitive Y chromosome of papaya. Genome Res. 18, 1938–1943. https://doi.org/10.1101/gr.078808.108 (2008).
Bergero, R., Qiu, S. & Charlesworth, D. Gene loss from a plant sex chromosome system. Curr. Biol. 25, 1234–1240. https://doi.org/10.1016/j.cub.2015.03.015 (2015).
Ming, R., Bendahmane, A. & Renner, S. S. Sex chromosomes in land plants. Annu. Rev. Plant Biol. 62, 485–514. https://doi.org/10.1146/annurev-arplant-042110-103914 (2011).
Charlesworth, D., Charlesworth, B. & Marais, G. Steps in the evolution of heteromorphic sex chromosomes. Heredity 95, 118–128. https://doi.org/10.1038/sj.hdy.6800697 (2005).
Bhattacharya, J. & Khuspe, S. S. In vitro and in vivo germination of papaya (Carica papaya L.) seeds. Sci. Hortic. 91, 39–49. https://doi.org/10.1016/S0304-4238(01)00237-0 (2001).
Sattler, M. C., Soares, F. A. F., Silva, J. C., Carvalho, C. R. & Clarindo, W. R. Physical mapping of 5S rDNA in Eucalyptus dunnii Maiden and Zea mays L. by PRINS. Cytologia 84, 77–83. https://doi.org/10.1508/cytologia.84.77 (2019).
Silva, J. C., Soares, F. A. F., Sattler, M. C. & Clarindo, W. R. Repetitive sequences and structural chromosome alterations promote intraspecific variations in Zea mays L. karyotype. Sci. Rep. 10, 1–9. https://doi.org/10.1038/s41598-020-65779-3 (2020).
Contim, L. A. S. et al. The soybean sucrose binding protein gene family: genomic organization, gene copy number and tissue-specific expression of the SBP2 promoter. J. Exp. Bot. 54, 2643–2653. https://doi.org/10.1093/jxb/erg301 (2003).
Silva, J. C., Carvalho, C. R. & Clarindo, W. R. Updating the maize karyotype by chromosome DNA sizing. PLoS One. 13, e0190428. https://doi.org/10.1371/journal.pone.0190428 (2018).
Soares, F. A. F. et al. Plant chromosome-specific probes by microdissection of a single chromosome: Is that a reality?. Front. Plant Sci. 11, 1–9. https://doi.org/10.3389/fpls.2020.00334 (2020).
Quadros, I. P. S. et al. Cadmium-mediated toxicity in plant cells is associated with the DCD/NRP-mediated cell death response. Plant Cell Environ. 1, 1–16. https://doi.org/10.1111/pce.14218 (2021).
Acknowledgements
We would like to thank the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brazil (CAPES)—Finance Code 001.
Author information
Authors and Affiliations
Contributions
The authors D.A.F and C.W.R. designed this work, conducted all experiments and wrote the manuscript. D.A.F., S.F.A.F., S.J.C. and C.W.R. conducted the in situ hybridization experiments. D.A.F. and S.M.C. conducted the sequencing bioinformatics. All authors equally contributed for manuscript editing and revision and approved the final manuscript for submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Dornela, A.F., Soares, F.A.F., Silva, J.C. et al. Carica papaya L. sex chromosome review and physical mapping of the serk 2, svp-like and mdar 4 sequences. Sci Rep 14, 14830 (2024). https://doi.org/10.1038/s41598-024-65880-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-65880-x
- Springer Nature Limited