Abstract
Carbapenem-resistant Enterobacteriaceae (CRE) have diminished treatment options causing serious morbidities and mortalities. This systematic review and meta-analysis assessed the prevalence and associated factors of Enterobacteriaceae infections in clinical, livestock and environmental settings globally. The population intervention comparison and outcome strategy was used to enroll studies using the preferred reporting system for systematic review and meta-analysis to include only cross-sectional studies. Search engines used to retrieve articles included journal author name estimator, PubMed, Google Scholar and African Journals Online (AJOL). The Newcastle–Ottawa scale was used to assess the quality of studies. Sixteen articles from 2013 to 2023 in Africa, Asia, Europe and South America were studied. The pooled prevalence of CRE was 43.06% (95% CI 21.57–66.03). Klebsiella pneumoniae (49.40%), Escherichia coli (26.42%), and Enterobacter cloacae (14.24%) were predominant. Klebsiella pneumoniae had the highest resistance with the blaKPC-2 in addition to blaNDM, blaOXA-48, blaIMP and blaVIM. The blaKPC-2 genes occurrence was associated with environmental (P-value < 0.0001) and South American studies (P-value < 0.0001), but there was no difference in the trends over time (P-value = 0.745). This study highlights the high rates of CRE infections, particularly within blaKPC production. Monitoring and surveillance programs, research and infection control measures should be strengthened. Additionally, further studies are needed to explore the mechanisms driving the predominance of specific bacterial species and the distribution of resistance genes within this bacterial family.
Similar content being viewed by others
Introduction
Enterobacteriaceae are a large and diverse group of gram-negative bacteria that are found in the environment and in the gut of animals, birds and humans. They can cause a variety of infections, including urinary tract infections, pneumonia, and sepsis. In recent years, there has been an increasing prevalence of Enterobacteriaceae that are resistant to several antibiotics1.
Carbapenem-resistant Enterobacteriaceae (CRE) are a group of Gram-negative bacteria that have become resistant to carbapenems, which are a class of antibiotics that are last-line treatments for many serious infections. The CRE can be found in a variety of settings, including hospitals, nursing homes and several other healthcare facilities1. However, they are also increasingly being found in the environment2,3,4 and among people who have or not been recently hospitalized and livestock5,6,7,8. The emergence of CRE in these various settings, has raised concerns about the widespread dissemination of these multidrug-resistant pathogens9,10. Various carbapenamases have been detected in Enterobacteriaceae organisms, categorized into three classes of β-lactamases: Ambler class-A (KPC, SME, IMI, NMC, GES types), class-B metallo-ß-lactamases (VIM, IMP, GIM, SMP, and NDM types), and class-D β-lactamases (primarily OXA-48). Additionally, there are rare chromosome-encoded class C enzymes. These enzyme groups all provide resistance to broad-spectrum antibiotics, including carbapenems11. Transfer of these resistance genes facilitated by processes such as transformation, transduction, and conjugation, is a common occurrence in Carbapenem-Resistant Enterobacteriaceae (CRE)11.
Several genes, including blaOXA-58, blaOXA-48, blaOXA-23, blaVIM-2, and blaVIM-4, were identified as playing a role in carbapenem resistance12. These genes were found to be commonly present in carbapenemase-producing bacteria during outbreaks in Africa by 201012. Carbapenemase-encoding genes, blaVIM, blaOXA, blaIMP, blaKPC, and blaNDM, showed variable distribution patterns among clinical isolates collected from Mulago National Referral Hospital in Uganda13. Additionally, bacterial strains producing Carbapenemase and carrying blaVIM and blaOXA-48 genes were also detected in specimens from patients at Mbarara Regional Referral Hospital14.
Elsewhere, these organisms have also been found in animals raised for food due to the excessive use of antibiotics in animal farming. The presence of CRE in livestock raises concerns about the potential transmission of antibiotic-resistant bacteria to humans. If these bacteria are able to move from animals to humans, it creates a significant risk to public health by reducing the effectiveness of treatment for severe infections15,16. The detection of CRE in both human and animal samples indicates that the environmental sources in rural southwestern Uganda could be aiding in the dissemination of resistant bacteria17. Therefore, it is essential to identify these sources and enforce necessary control measures17.
Therefore, there is a need for collective efforts globally to control AMR including CRE. The One Health concept, which emphasizes the interconnectedness of human, animal, and environmental health, is crucial for tackling the global issue of CRE prevalence. Collaborative efforts across these domains are essential for comprehensive control measures. Understanding the genetic constitution of the CRE would serve the primary role of unlocking this burden.
Aim of the study
The aim of this systematic review and meta-analysis was to assess the prevalence and transmission dynamics of Enterobacteriaceae in clinical, livestock, and environmental settings globally.
Methods
Selection
The population intervention comparison and outcome (PICO) strategy was used to make selection of the studies. Only cross-sectional studies that employed the molecular assays to assess CRE during data collection were included in the review. They should have been published in English. For clinical studies, those that included stool specimens were excluded in the review. Comparative groups were clinical, environmental and livestock with prevalence as the measure of outcome.
Data search
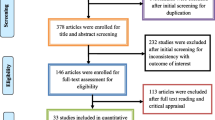
The Preferred Reporting System for Systematic Review and Meta-analysis (PRISMA) standards was used to guide the process that includes literature search and data extraction, data cleaning, data analysis and interpreting result a seen in Fig. 1. The primary studies that investigated epidemiology, transmission dynamics of molecular analysis of carbapenem resistant Enterobacteriaceae in community and health care settings were adopted for the systematic review. Search engines used to retrieve the articles included journal author name estimator (JANE), PubMed, Google scholar and African Journals Online (AJOL). Data from articles published between January 2010 and December 2022 was included.
Search terms were (“Carbapenem Resistant Enterobacteriaceae”), (“Hospital Carbapenem Resistant Enterobacteriaceae”), (“Community Carbapenem Resistant Enterobacteriaceae”), (“Carbapenem Resistant Enterobacteriaceae in livestock”), (“Environment Carbapenem Resistant Enterobacteriaceae”) and, from the articles databases. The (“Phylogeny relationship of Carbapenem Resistant Enterobacteriaceae”) and (“Nucleotide homology comparison of Carbapenem Resistant Enterobacteriaceae”) search term were used to extract data from the DNA databases. Snow balling was done after text review of references for related articles. Search articles were organized in Endnote from which the duplicates were removed.
Screening
A thorough screening of the articles was performed starting with title and abstract and then full text reading. This was performed by two reviewers (Ms. Barbra Tuhamize and Assoc Prof. Joel Bazira) from 1st August to 21st October 2023. The two also performed quality appraisal of the recruited articles as cross-sectional in nature to avoid bias using the Cochrane risk of bias assessment tool. Any disagreement between the two reviewers were settled by a consensus during the weekly evaluation meetings that sat every Wednesday of the week.
Assessment of data quality
The Newcastle–Ottawa scale (NOS) was used to assess the quality of studies that passed the text reviews. Studies with a NOS of three or less were dropped from the meta-analysis.
The meta-analysis (quantitative data synthesis)
The effect size for each study was checked and later the combined estimate of effect size generated. The random effects model was considered to include heterogeneity. The forest plot was drawn to check the heterogeneity. The funnel plot and the Egger’s and Beggs tests were used to assess the publication bias. Sensitivity analysis was performed by removing one article at ago and check result variation.
Subgroup analysis was later performed for the highly predominant blaKPC gene amongst the different sample sources, continent and trends over time. The distribution of the blaKPC gene among the sample sources and the different continents was performed using the metanalysis of pooled prevalence using the chi square test at 95% CI and trends over time from 2013 to 2023 using linear regression at 95% CI.
Ethics approval and consent to participate
This is a systematic review and meta-analysis. We reviewed published studies and only those that had received ethics approval during data collection.
Results
Systematic review
The search and systematic review yielded 16 studies from 2013 to 2023 in Africa, Asia, Europe and South America (Table 1) using the PRISMA standards guide (Fig. 1).
These 16 studies that were included in the study, were caried out from African, Asia, Europe and South America using samples from environmental, livestock and clinical specimens1,2,3,4,5,6,7,8,9,10,13,18,19,20,21. Several Enterobacteriaceae were reported to carry resistance genes as shown in (Table 1) below.
A meta-analysis was conducted among the 16 articles. The pooled prevalence of CRE was 43.06% (95% confidence interval [CI] 21.57–66.03) using a random effect model, catering for heterogeneity (Fig. 2). Bias was assessed using the funnel plot indicated in (Fig. 3) below.
The predominant bacteria species were, Klebsiella pneumoniae (49.40%), Escherichia coli (26.42%), and Enterobacter cloacae (14.24%). Other bacteria species included Klebsiella oxytoca (2.99%), Proteus mirabilis (2.55%), Enterobacter aerogenes (1.95%), Proteus vulgaris (0.19%), Kluyvera (0.19%), Morganella morganii (0.19%), Citrobacter freundii (0.99%), Serratia marcescens (0.65%), Citrobacter braakii (0.19%), and Citrobacter koseri (0.05%), Providencia rettgeri (0.05%), Providencia stuartii (0.05%), Raoultella ornithinolytica (0.05%) were the least isolated (Table 2).
High prevalence of CRE was exhibited among Klebsiella pneumoniae isolates. The gene occurrence was higher for the blaKPC-2, followed by blaNDM, blaOXA-48, blaIMP, blaVIM, and the least was blaGES, no resistance was reported by Citrobacter braakii, Klebsiella oxytoca, Proteus mirabilis, Proteus vulgaris, and Providencia rettgeri (Table 3). Organisms carried resistance genes from one to as many as seven per bacteria.
The blaKPC-2 gene was dominant, carried more among Klebsiella pneumoniae (582/989; 59%), followed by Kluyvera spp (2/4; 50%), Serratia marcescens (3/13; 23%) and lowly carried by Enterobacter cloacae (14/285; 5%), Citrobacter freundii (1/20; 5%) and Escherichia coli (5/529; 1%) (Fig. 4).
The highly prevalent blaKPC gene’s occurrence was evaluated amongst the sample sources (Table 4), continents (Table 5) and trends over time from 2013 to 2023 (Fig. 5). Environmental samples recorded the highest prevalence of the blaKPC gene compared the humans (P-value < 0.0001) and livestock (P-value < 0.0001). Amongst the continents, South American studies produced the highest as compared to Asia (P-value < 0.0001), Africa (P-value < 0.0001) and Europe (P-value < 0.0001). There was no significant difference in the trends of occurrence of the blaKPC gene over time (P-value = 0.745).
Discussion
The findings of this systematic review and meta-analysis provide critical insights into the prevalence and characteristics of Enterobacteriaceae infections, as well as the distribution of carbapenemase resistance genes within this bacterial family. These bacteria can cause a wide range of infections, including urinary tract infections, surgical site infections, pneumonia, bloodstream infections, and diarrhea22,23,24. Enterobacteriaceae infections can be particularly serious in young children, the elderly, and people with weakened immune systems23. The results from this systematic review and meta-analysis have important implications for public health to curb AMR.
Prevalence of Enterobacteriaceae infections
This systematic review and meta-analysis were unique that they pooled studies that researched on several sample sources including humans (urine samples), livestock (feces and cloaca) and in the environment.
The findings of this meta-analysis, based on a comprehensive review of 16 articles, provide important insights into the global prevalence of Carbapenem-resistant Enterobacteriaceae infections. The pooled prevalence of CRE infections was estimated at 43.06%, (95% CI 21.57–66.03%). Notably, a random effect model was employed to account for the observed heterogeneity among the studies. This finding is consistent with the increasing reports of CRE infections from various parts of the world, reflecting the growing challenge of antibiotic resistance in clinical and non-clinical settings25. The observed pooled prevalence of 43.06% underscores the alarming global prevalence of CRE infections, suggesting that this issue is more widespread than previously thought.
The wide 95% confidence interval (21.57–66.03%) reflects the substantial variability in CRE prevalence across different regions and populations. The heterogeneity observed in the meta-analysis may be attributed to several factors, including variations in healthcare practices, antibiotic use, and infection control measures among different countries and healthcare settings26. This heterogeneity highlights the need for a multifaceted approach to address CRE, as a one-size-fits-all strategy may not be sufficient.
It is essential to contextualize the implications of this prevalence estimate in the broader landscape of antimicrobial resistance. CRE is particularly concerning due to their resistance to carbapenem antibiotics, which are often considered the last line of defense against severe infections caused by Gram-negative bacteria24. The high prevalence of CRE signifies a critical need for effective interventions to curb the spread of resistant strains and preserve the efficacy of existing antibiotics. Like Kelly et al.25 suggested, this is one area prime to support especially through the one health approach if AMR must be tamed.
In addition to the overall prevalence estimate, the findings of this meta-analysis provide a foundation for understanding the scope of the CRE problem across various populations and regions. This information is instrumental in guiding public health policies, infection control strategies, and antimicrobial stewardship programs aimed at mitigating the impact of CRE.
To address the challenges posed by CRE, we concur with27 that a comprehensive approach is warranted, including the judicious use of antibiotics, stringent infection control measures in healthcare facilities, and active surveillance to detect and contain CRE outbreaks. Furthermore, international collaboration is essential to share best practices and prevent the further dissemination of CRE strains.
It is important to note that the limitations of this meta-analysis include potential publication bias and variations in data reporting and methodology among the included studies. Future research should continue to monitor CRE prevalence and investigate the genetic mechanisms underlying carbapenem resistance to develop targeted interventions.
Predominant bacterial species
The identification of predominant bacterial species within the Enterobacteriaceae family is essential for understanding the epidemiology of these infections. Klebsiella pneumoniae emerged as the most prevalent species, accounting for 49.40% of isolates followed by Escherichia coli and Enterobacter cloacae with prevalence rates of 26.42% and 14.24%, respectively.
The predominance of K. pneumoniae would be explained by their isolation from clinical studies abundantly, whose (the clinical samples) sample sizes were higher than those from the environment and livestock. This distribution of predominant species suggests that certain Enterobacteriaceae members may possess specific virulence factors or resistance mechanisms that contribute to their prevalence. Some known highly virulent bacteria including Salmonella and Shigella were not reported, but this is explained by them being enteric (by transmission) and very rarely or occassionally cause urinary tract infections28.
The abundance of Enterobacteriaceae in environmental sources is an indicator of their animal and human fecal sources29, and the cross distribution of carbapenem resistance genes worries AMR control program and the One health approach2,30,31. Moreover, Kluyvera, a less human pathogen (reference) was found to carry blaKPC genes (that is predominantly isolated from clinical specimens). This highlights a need to study comprehensively several other environmental sources such as indoor air and surface areas from public places including schools, hotels, office spaces and day care centers among others.
Further research is warranted to elucidate the factors driving the predominance of these species and to develop targeted interventions for their control.
Carbapenemase resistance genes
One of the most concerning aspects of Enterobacteriaceae infections is the emergence and spread of carbapenem-resistant strains (CRE). Our analysis identified Klebsiella pneumoniae as a major contributor to CRE prevalence, with a notably high occurrence of carbapenemase resistance genes within this species.
Among the carbapenemase resistance genes, blaKPC-2 exhibited the highest occurrence, followed by blaNDM, blaOXA-48, blaIMP, and blaVIM being the least prevalent. The diversity of carbapenemase genes highlights the complexity of antimicrobial resistance within the Enterobacteriaceae family. In environmental and sewage sources, several authors have reported about the abundance of the CRE and their potential to dissemination8,32,33,34,35. Therefore, the presence of these genes underscores the urgent need for stringent infection control measures and the judicious use of antibiotics to prevent further dissemination of CRE33.
It is worth noting that certain bacterial species, including Proteus mirabilis (51) and Klebsiella oxytoca (60) never carried any resistance genes. These species could serve as valuable models for studying resistance mechanisms and potentially inform strategies for combating CRE. However, a similar observation cannot be made for Raoultella ornithinolytica, Proteus vulgaris, Citrobacter braakii, Citrobacter koseri, Providencia rettgeri, Providencia stuartii and Morganella morganii for not expressing any resistance genes because their frequencies were too low to generate any statistical inference.
The blaKPC dominance
The findings of this systematic review and meta-analysis reveal crucial insights into the prevalence and distribution of the blaKPC-2 gene among different bacterial species in diverse settings, shedding light on the genetic determinants of Carbapenem-resistant Enterobacteriaceae (CRE).
The dominance of the blaKPC-2 gene, particularly in Klebsiella pneumoniae, is a significant and concerning finding. Klebsiella pneumoniae is a well-known nosocomial pathogen responsible for a wide range of infections, and its association with the blaKPC-2 gene indicates the potential for challenging therapeutic outcomes36. The high prevalence of this gene among Klebsiella pneumoniae (59%) is similar to what37 reported in China. This underscores the importance of this species as a key contributor to the CRE problem, especially in healthcare settings where carbapenems are frequently used.
Furthermore, the detection of the blaKPC-2 gene in other species like Kluyvera spp, Serratia marcescens, Enterobacter cloacae, Citrobacter freundii, and Escherichia coli highlights the diversity of bacterial species capable of carrying this resistance gene. While these species might not be as common as Klebsiella pneumoniae in clinical infections, their capacity to harbor blaKPC-2 is a critical concern. In particular, the presence of this gene in Kluyvera spp and Serratia marcescens (which are commonly found in the environment) is noteworthy, given the potential for these species to act as reservoirs for resistance genes and share them with more clinically relevant pathogens.
The variation in the prevalence of the blaKPC-2 gene across different bacterial species suggests that the distribution of carbapenem resistance determinants is not uniform and may be influenced by species-specific factors and selective pressures. For example, Enterobacter cloacae and Escherichia coli exhibit lower prevalences (5% and 1%, respectively) of the blaKPC-2 gene, indicating that other resistance mechanisms may be more prevalent in these species or that their association with CRE may be mediated by different resistance genes encoded by plasmids or bacteriophages38. These variations are consistent with previous studies that have reported differential resistance gene carriage among different Enterobacteriaceae species39.
The identification and prevalence of specific carbapenemase genes, such as the blaKPC-2 gene, within CRE are critical for understanding the dynamics of resistance gene dissemination across different reservoirs, including environmental samples, humans, and livestock. This systematic review and meta-analysis have revealed noteworthy findings regarding the dominance and prevalence of the blaKPC-2 gene in these diverse reservoirs.
Understanding the diversity of species carrying the blaKPC-2 gene is crucial for the development of effective infection control and antimicrobial stewardship strategies. Also, to focusing on well-known CRE pathogens like Klebsiella pneumoniae, efforts should also target species with lower prevalence but potential for gene dissemination. Additionally, monitoring and surveillance programs should consider the presence of the blaKPC-2 gene in environmental samples, as these reservoirs can contribute to the spread of resistance determinants among both clinical and non-clinical settings.
The findings of this study emphasize the importance of continued research into the genetic determinants of CRE and the factors influencing their distribution among different bacterial species. By understanding the prevalence of resistance genes like blaKPC-2, we can better address the evolving threat of CRE and develop strategies to mitigate its impact on public health.
The study by3 only reported blaNMD and blaOXA but not blaIMP, blaKPC, blaVIM and blaGES similar to. This was different from other studies that reported at least one of these genes with dominance of blaKPC.
Attributes to blaKPC occurrence
The ecological niche of the blaKPC gene
In this research, a higher prevalence of the blaKPC gene in environmental samples was obtained compared to both humans and livestock. This result suggests that environmental sources, such as water, soil, or wastewater, may serve as a reservoir for CRE carrying the blaKPC gene especially in community transmission. Several factors could contribute to this higher prevalence in environmental though none of the studies assessed these factors. The environment can act as a bridge for CRE transmission between humans, animals, and the surrounding ecosystem1. The contamination of the environment with CRE from human or livestock sources can establish a cycle of transmission.
Understanding the higher prevalence of the blaKPC gene in environmental samples emphasizes the need for improved environmental surveillance and management to reduce the dissemination of carbapenem resistance. It is essential to consider the complex interplay between human activities, the environment, and antimicrobial resistance.
These findings underscore the importance of adopting a “One Health” approach, which recognizes the interconnectedness of human, animal, and environmental health. Efforts should be made to monitor and mitigate CRE in all these domains to effectively combat the global spread of antibiotic resistance.
The systematic review and meta-analysis of CRE prevalence and genetic determinants, particularly the blaKPC-2 gene, have yielded intriguing insights into the distribution of carbapenem resistance across continents. The findings not only highlight the dominance of the blaKPC-2 gene but also reveal significant variations in its prevalence among different continents.
Variation in prevalence among continents
The marked differences in blaKPC gene prevalence among continents are of substantial concern and indicate significant geographical disparities in CRE distribution:
The highest prevalence of the blaKPC gene in South American studies, as compared to Asia, Africa, and Europe, points to a particularly critical situation in this region. South America has witnessed increasing challenges in healthcare-associated infections and antimicrobial resistance, which may be contributing to the high prevalence of CRE.
Asia came second to South America and this is similar to recent reports. Asia has been a significant focus of CRE research due to its large population and diverse healthcare systems. Various studies have highlighted the presence of blaKPC-carrying CRE in the region, indicating the multifaceted nature of the issue.
The comparatively lower prevalence of the blaKPC gene in Africa may be influenced by variations in healthcare infrastructure, antimicrobial use, and epidemiological factors. However, CRE has been a growing concern in parts of Africa, and its prevalence is expected to evolve.
Europe's comparatively lower prevalence suggests a more controlled situation, likely due to stringent infection control measures and antibiotic stewardship programs in place. However, localized outbreaks and variations within European countries are also evident.
Consistent trends in blaKPC gene occurrence over time
The systematic review and meta-analysis findings, highlighting the dominance of the blaKPC-2 gene, and the consistent trends in the occurrence of the blaKPC gene over a ten-year period (2013–2023), provide valuable insights into the dynamics of carbapenem resistance in CRE. These findings indicate the persistence and resilience of this carbapenemase gene within the global CRE landscape.
It is important to note that while the overall trends in the occurrence of the blaKPC gene appear stable, there may be localized variations and fluctuations within different regions, healthcare facilities, and ecological niches. These nuances could be influenced by factors such as local infection control practices, antibiotic use patterns, and the introduction of new CRE strains25,40. The observed stability in the occurrence of the blaKPC gene from 2013 to 2023 reflects the ongoing challenges posed by carbapenem resistance. These results emphasize the importance of continued vigilance, surveillance, and the development of strategies to combat CRE and its associated carbapenemase genes.
Limitations
While this systematic review and meta-analysis provide valuable insights, some limitations should be acknowledged. There may be heterogeneity among the included studies, which could impact the generalizability of the results. The analysis is based on existing literature up to the cutoff date and may not capture more recent developments in the field. The risk factors to CRE were not presented for all the studies that qualified to the meta-analysis which makes it difficult to establish control measures. Publication bias could affect the prevalence estimates, as studies with significant findings are more likely to be published.
Implications for public health
The findings of this study have important implications for public health. The high prevalence of Enterobacteriaceae infections, especially in hospitals, highlights the need for infection prevention and control measures. These measures should include hand hygiene, proper cleaning and disinfection of surfaces, and the use of antibiotics only when necessary.
The increasing prevalence of Carbapenem-resistant Enterobacteriaceae is a serious threat to public health. These bacteria can be difficult to treat, and they can cause serious infections that can lead to death. It is important to continue research on new antibiotics and other treatments for Enterobacteriaceae infections.
There is need to improve article scrutiny during review to allow better information generation that could potentially lead to control of the MDR bacteria isolates.
Conclusion
In conclusion, this systematic review and meta-analysis highlight the substantial prevalence (43.06% (95% CI 21.57–66.03)) of Enterobacteriaceae infections, the variability in prevalence based on the source of isolation, and the concerning emergence of carbapenem-resistant strains, particularly within Klebsiella pneumoniae, particularly the blaKPC production. Monitoring and surveillance programs should consider the presence of the blaKPC-2 gene in environmental samples, as these reservoirs can contribute to the spread of resistance determinants among both clinical and non-clinical settings. These findings emphasize the importance of ongoing research, surveillance, and infection control measures to address the public health challenges posed by Enterobacteriaceae infections and antimicrobial resistance. Additionally, further studies are needed to explore the mechanisms driving the predominance of specific bacterial species and the distribution of resistance genes within this bacterial family.
Recommendations
In clinical practice, the utilization of carbapenems should strictly follow resistance testing methods such as both DST and PCR. Additionally, when administering carbapenems to individuals, it is recommended to combine them with carbapenemase inhibitors as a proactive measure. It is crucial to conduct comprehensive large-scale studies focusing on the prevalence of blaKPC, blaNDM, blaOXA-48, blaIMP and blaVIM in Enterobacteriaceae to comprehend the spectrum of resistant genes in both community and healthcare environments.
Data availability
Data is available with journal author name estimator (JANE), PubMed, Google scholar and African Journals Online (AJOL).
References
Jin, L. et al. Emergence of mcr-1 and carbapenemase genes in hospital sewage water in Beijing, China. J. Antimicrob. Chemother. 73(1), 84–87. https://doi.org/10.1093/jac/dkx355 (2017).
Montezzi, L. F. et al. Occurrence of carbapenemase-producing bacteria in coastal recreational waters. Int. J. Antimicrob. Agents 45(2), 174–177 (2015).
Su, Y. et al. Drug resistance analysis of three types of avian-origin carbapenem-resistant Enterobacteriaceae in Shandong Province, China. Poult. Sci. 102(3), 102483. https://doi.org/10.1016/j.psj.2023.102483 (2023).
Piedra-Carrasco, N. et al. Carbapenemase-producing Enterobacteriaceae recovered from a Spanish river ecosystem. PLoS One 12(4), e0175246 (2017).
Makharita, R. R. et al. Antibiogram and genetic characterization of carbapenem-resistant gram-negative pathogens incriminated in healthcare-associated infections. Infect. Drug Resist. 13, 3991–4002. https://doi.org/10.2147/IDR.S276975 (2020).
Nabti, L. Z. et al. Molecular characterization of clinical carbapenem-resistant Enterobacteriaceae isolates from Sétif, Algeria. Microb. Drug Resist. 28(3), 274–279. https://doi.org/10.1089/mdr.2021.0123 (2022).
Cizmeci, Z., Aktas, E., Otlu, B., Acikgoz, O. & Ordekci, S. Molecular characterization of carbapenem-resistant Enterobacteriaceae yields increasing rates of NDM-1 carbapenemases and colistin resistance in an OXA-48-endemic area. J. Chemother. 29(6), 344–350. https://doi.org/10.1080/1120009x.2017.1323149 (2017).
Feng, J. et al. Characterization of carbapenem-resistant Enterobacteriaceae cultured from retail meat products, patients, and porcine excrement in China. Front. Microbiol. 12, 743468. https://doi.org/10.3389/fmicb.2021.743468 (2021).
Chabou, S., Leulmi, H., Davoust, B., Aouadi, A. & Rolain, J.-M. Prevalence of extended-spectrum β-lactamase- and carbapenemase-encoding genes in poultry faeces from Algeria and Marseille, France. J. Glob. Antimicrob. Resist. 13, 28–32. https://doi.org/10.1016/j.jgar.2017.11.002 (2018).
Abdallah, H. M. et al. Extended-spectrum β-lactamases and/or carbapenemases-producing Enterobacteriaceae isolated from retail chicken meat in Zagazig, Egypt. PLoS One 10(8), e0136052. https://doi.org/10.1371/journal.pone.0136052 (2015).
Nordmann, P., Dortet, L. & Poirel, L. Carbapenem resistance in Enterobacteriaceae: Here is the storm!. Trends Mol. Med. 18(5), 263–272 (2012).
Kateete, D. P. et al. Carbapenem resistant Pseudomonas aeruginosa and Acinetobacter baumannii at Mulago hospital in Kampala, Uganda (2007–2009). Springerplus 5, 1–11 (2016).
Okoche, D., Asiimwe, B. B., Katabazi, F. A., Kato, L. & Najjuka, C. F. Prevalence and characterization of carbapenem-resistant Enterobacteriaceae isolated from Mulago National Referral Hospital, Uganda. PloS One 10(8), e0135745 (2015).
Ampaire, L. M., Katawera, V., Nyehangane, D., Boum, Y. & Bazira, J. Epidemiology of carbapenem resistance among multi-drug resistant Enterobacteriaceae in Uganda. Br. Microbiol. Res. J. 8(2), 418 (2015).
Muloi, D. et al. A cross-sectional survey of practices and knowledge among antibiotic retailers in Nairobi, Kenya. J. Glob. Health 9(2), 020412 (2019).
Murray, C. J. et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. The Lancet 399(10325), 629–655 (2022).
Tuhamize, B. et al. Klebsiella pneumoniae carbapenamases in Escherichia coli isolated from humans and livestock in rural south-western Uganda. Plos One 18(7), e0288243 (2023).
Ribeiro, V. B. et al. Molecular characterization of Klebsiella pneumoniae carbapenemase-producing isolates in southern Brazil. J. Med. Microbiol. 62(11), 1721–1727. https://doi.org/10.1099/jmm.0.062141-0 (2013).
Li, Y. et al. Rapid increase in prevalence of carbapenem-resistant Enterobacteriaceae (CRE) and emergence of colistin resistance gene mcr-1 in CRE in a hospital in Henan, China. J. Clin. Microbiol. 56(4), 10–1128. https://doi.org/10.1128/jcm.01932-17 (2018).
Solgi, H. et al. Molecular characterization of intestinal carriage of carbapenem-resistant Enterobacteriaceae among inpatients at two Iranian university hospitals: First report of co-production of blaNDM-7 and blaOXA-48. Eur. J. Clin. Microbiol. Infect. Dis. 36(11), 2127–2135. https://doi.org/10.1007/s10096-017-3035-3 (2017).
Sonnevend, Á. et al. Characterization of carbapenem-resistant Enterobacteriaceae with high rate of autochthonous transmission in the Arabian peninsula. PLoS One 10(6), e0131372. https://doi.org/10.1371/journal.pone.0131372 (2015).
Klein, R. D. & Hultgren, S. J. Urinary tract infections: microbial pathogenesis, host-pathogen interactions and new treatment strategies. Nat. Rev. Microbiol. 18(4), 211–226. https://doi.org/10.1038/s41579-020-0324-0 (2020).
Kłos, M. & Wójkowska-Mach, J. Hospital-acquired Enterobacteriaceae bloodstream infections in children. Dev. Period Med. 23(2), 131–136. https://doi.org/10.34763/devperiodmed.20192302.131136 (2019).
Darlene, M. & Anne, B. Enterobacteriaceae, Vol. 1. [Online]. Available: https://text.apic.org/toc/healthcare-associated-pathogens-and-diseases/enterobacteriaceae.
Kelly, A. M., Mathema, B. & Larson, E. L. Carbapenem-resistant Enterobacteriaceae in the community: A scoping review. Int. J. Antimicrob. Agents 50(2), 127–134. https://doi.org/10.1016/j.ijantimicag.2017.03.012 (2017).
van Loon, K., Voor In’t Holt, A. F. & Vos, M. C. A systematic review and meta-analyses of the clinical epidemiology of carbapenem-resistant Enterobacteriaceae. Antimicrob. Agents Chemother. 62(1), 10–1128. https://doi.org/10.1128/aac.01730-17 (2018).
Woodford, N., Wareham, D. W., Guerra, B. & Teale, C. Carbapenemase-producing Enterobacteriaceae and non-Enterobacteriaceae from animals and the environment: An emerging public health risk of our own making?. J. Antimicrob. Chemother. 69(2), 287–291. https://doi.org/10.1093/jac/dkt392 (2013).
MayoClinic. Shigella infection. Disease and conditions. [Online]. Available: https://www.mayoclinic.org/diseases-conditions/shigella/symptoms-causes/syc-20377529.
Davies, J. & Davies, D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 74(3), 417–433 (2010).
Pavez, M. et al. Molecular mechanisms of membrane impermeability in clinical isolates of Enterobacteriaceae exposed to imipenem selective pressure. Int. J. Antimicrob. Agents 48(1), 78–85 (2016).
Paschoal, R. P., Campana, E. H., Castro, Ld. S. & Picao, R. C. Predictors of carbapenemase-producing bacteria occurrence in polluted coastal waters. Environ. Pollut. 264, 114776 (2020).
Szekeres, E. et al. Abundance of antibiotics, antibiotic resistance genes and bacterial community composition in wastewater effluents from different Romanian hospitals. Environ. Pollut. 225, 304–315. https://doi.org/10.1016/j.envpol.2017.01.054 (2017).
Li, H., Cheng, W., Li, B., Xu, Y. & Zheng, X. The fate of antibiotic resistance genes during co-composting of swine manure with cauliflower and corn straw. Bioresour. Technol. 300, 122669. https://doi.org/10.1016/j.biortech.2019.122669 (2020).
Haller, L. et al. Occurrence and characteristics of extended-spectrum β-lactamase- and carbapenemase-producing bacteria from hospital effluents in Singapore. Sci. Total Environ. 615, 1119–1125. https://doi.org/10.1016/j.scitotenv.2017.09.217 (2018).
Gong, X. et al. Molecular characterization and epidemiology of carbapenem non-susceptible Enterobacteriaceae isolated from the Eastern region of Heilongjiang Province, China. BMC Infect. Dis. 18(1), 417. https://doi.org/10.1186/s12879-018-3294-3 (2018).
CDC. Klebsiella pneumoniae in Healthcare Settings. [Online]. Available: https://www.cdc.gov/hai/organisms/klebsiella/klebsiella.html#:~:text=When%20bacteria%20such%20as%20Klebsiella%20pneumoniae%20produce%20an,to%20kill%20the%20bacteria%20and%20treat%20the%20infection.
Su, C. et al. High prevalence of Klebsiella pneumoniae infections in AnHui province: Clinical characteristic and antimicrobial resistance. Infect. Drug Resist. 14, 5069–5078. https://doi.org/10.2147/idr.S336451 (2021).
Liu, H. et al. Distribution of β-lactamase genes and genetic context of blaKPC-2 in clinical carbapenemase-producing Klebsiella pneumoniae isolates. Infect. Drug Resist. 14, 237–247. https://doi.org/10.2147/idr.S290434 (2021).
Partridge, S. R. Resistance mechanisms in Enterobacteriaceae. Pathology 47(3), 276–284. https://doi.org/10.1097/pat.0000000000000237 (2015).
Tilahun, M., Kassa, Y., Gedefie, A. & Ashagire, M. Emerging carbapenem-resistant Enterobacteriaceae infection, its epidemiology and novel treatment options: A review. Infect. Drug Resist. 14, 4363–4374. https://doi.org/10.2147/idr.S337611 (2021).
Hamza, E. et al. Carbapenemase-producing Klebsiella pneumoniae in broiler poultry farming in Egypt. J. Glob. Antimicrob. Resist. 7, 8–10 (2016).
Alraddadi, B. M. et al. Molecular epidemiology and outcome of carbapenem-resistant Enterobacterales in Saudi Arabia. BMC Infect. Dis. 22(1), 542 (2022).
Acknowledgements
The department of microbiology faculty of medicine, Mbarara University of Science and Technology. Authors of the primary articles including; Solgi et al.20, Makharita et al.5, Ribeiro et al.18, Nabti et al.6, Cizmeci et al.7, Li et al.19, Hamza et al.41, Sonnevend et al.21, Jin et al.1, Montezzi et al.2, Okoche et al.13, Su et al.3, Alraddadi et al.42, Feng et al.8, Abdallah et al.10 and Piedra-Carrasco et al.4.
Author information
Authors and Affiliations
Contributions
Both authors contributed equally in data extraction, review, analysis and discussion. We have referenced all articles reviewed.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tuhamize, B., Bazira, J. Carbapenem-resistant Enterobacteriaceae in the livestock, humans and environmental samples around the globe: a systematic review and meta-analysis. Sci Rep 14, 16333 (2024). https://doi.org/10.1038/s41598-024-64992-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-64992-8
- Springer Nature Limited