Abstract
FAT atypical cadherin 1 (FAT1) is one of the most mutagenic genes in tumors, and several critical studies have revealed its role in tumors, although no pan-cancer studies are currently available. Therefore, we explored the potential oncogenic role of FAT1 in 33 tumors based on The Cancer Genome Atlas and Gene Expression Omibus datasets. We found that FAT1 was strongly expressed in most tumors and significantly correlated with their prognosis. Additionally, we analyzed the association of FAT1 with tumors from multiple perspectives, including single-cell sequencing, mutations, high tumor mutational burden, microsatellite instability, immune cell infiltration, and immune microenvironment. Our first pan-cancer study provided a relatively comprehensive understanding of the oncogenic role of FAT1 in tumors.
Similar content being viewed by others
Introduction
The recent article published in Nature1 revealed that FAT atypical cadherin 1 (FAT1) mutations play critical roles in tumors and, combined with a literature analysis, highlighted its multiple functions in different tumors. At this stage, performing pan-cancer analysis of significant genes in combination with high-volume tumor sequencing data was crucial and feasible.
The public database, TCGA project2,3, and GEO database4, containing functional genomic datasets of different tumors, enabled us to perform a pan-cancer analysis. FAT1 encodes a protocadherin, which is one of the most frequently mutated genes in human cancers and plays various functions1,5,6. However, there was no evidence of its relationship with multiple tumor types.
Our study was the first pan-cancer analysis of FAT1 using the TCGA and GEO databases. We explored the potential molecular mechanisms of FAT1 in different tumorigenesis and clinical prognosis in terms of gene expression, survival status, genetic alterations, protein phosphorylation, immune infiltration, associated cellular pathways, and marker gene relevance.
Materials and methods
Study design
Our study was deemed exempt from institutional board approval and patient informed consent because all data were deidentified and publicly available. This study analyzed the multiple features of FAT1 in the cancer genome atlas pan-cancer cohort to pave the road for in-depth experimental studies. We confirmed that all methods were carried out following relevant guidelines and regulations.
Data and tools
The TCGA (https://portal.gdc.cancer.gov/) database contained sequencing and clinicopathological data from patients with over 30 different types of tumors. Sequencing data of 33 tumors were downloaded from the TCGA database (10,327 tumor samples and 730 matched paraneoplastic samples) (Table 1).
For some tumors without normal or highly restricted normal tissues, their expression data from the GTEx database were obtained using GEPIA2 (http://gepia2.cancer-pku.cn/) webserver7. Additionally, survival duration information of 10,327 tumor samples was also downloaded from the TCGA database (including OS, DSS, DFI, and PFI). The gene mutation data of 10,114 pan-cancer samples and the MSI score information of 10,415 tumor samples were downloaded from the TCGA database. The pan-cancer samples and analytical tools included in Sangerbox 3.0 were used in this study. This study mainly used R version 3.6.1 software (https://www.r-project.org/) and various practical packages to process and visualize the data. Other web servers used are introduced separately in different sections throughout the manuscript. All data used in this study were standardly cited as per the requirements of public databases and were permitted for use.
Differential expression analysis
The mRNA differential expression of FAT1 in pan-cancer was analyzed using the ggpubr package and Wilcox test. For tumors lacking paracancerous samples and insufficient sample size (ACC, CESC, DLBC, ESCA, GBM, LAML, LGG, OV, PAAD, SKCM, TGCT, and UCS), the differential expression box map of FAT1 in “Match TCGA normal and GTEx data” was searched in GEPIA2, with p-value Cutoff = 0.01, |Log2FC|Cutoff = 1. To confirm the reliability of our results, the network tool Sangerbox 3.0 (http://vip.sangerbox.com/) database was used to search the expression of FAT1 in pan-cancer. This tool is based on the unified and standardized pan-cancer data set of UCSC Genome Browser (http://genome-asia.ucsc.edu/): TCGA TARGET GTEx (PANCAN, N = 19,131, G = 60,499). The expression data of FAT1 were extracted, the samples with zero expression levels were filtered, and the expression values were converted using log2 (x + 0.001). The cancer species with less than three samples in a single cancer specie were excluded, and the difference was analyzed by unpaired Wilcoxon Rank Sum and Signed Rank Tests. Oncomine8 (https://www.oncomine.org/) tumor data set was also included to analyze the mRNA expression of FAT1. We combined data of FAT1 expression and tumor staging of patients, used the Wilcox test of the limma package for analysis, controlled the false discovery rate (FDR) below 0.05, and then plotted the graphs using ggpubr. We also searched the protein expression of FAT1, immunohistochemistry9, and single-cell RNA expression in UALCAN10 (http://ualcan.path.uab.edu/) and The Human Protein Atlas (https://www.proteinatlas.org/).
Survival prognosis analysis
We searched the relationship between FAT1 expression and pan-cancer patients prognosis using Sangerbox, which is based on the TCGA TARGET GTEx dataset. The TCGA prognosis study published in Cell journal11 provided a high-quality prognostic data set of TCGA. TARGET follow-up data were obtained from UCSC as supplementary data. Additionally, the samples with no expression level and follow-up duration of less than 30 days were excluded, and the expression data were transformed using log2 (x + 0.001). Additionally, the cancers with less than ten samples were also excluded, and the pan-cancer expression data and the OS, DSS, DFI, and PFI data of the corresponding samples were also retrieved. Cox proportional hazard regression model was established by the coxph function of survival (version 3.2–7) package to analyze the relationship between gene expression and prognosis in each tumor. The Kaplan–Meier method was used to determine the prognostic value of FAT1 based on the downloaded transcriptome and clinicopathological data. Log-rank test was used to obtain a significant prognosis.
Mutation analysis
The cBioPortal database (http://www.cbioportal.org/)12 is a web service that integrates the downloading of tumor datasets with online analysis and provides a variety of robust and reliable analysis tools. The mutations of FAT1 in pan-cancer (including mutations, structural variants, and copy number alterations) were analyzed based on the 32 studies selected (10,967 samples, TCGA, PanCancer Atlas). Additionally, all likely oncogenic mutations (OncoKB), Post Translational Modifications (PTMs), mutated exons, and details regarding the subcellular location of the mature FAT1 protein were analyzed. Gene mutations may affect the survival of tumor patients. To analyze the relationship between FAT1 mutations and patient prognosis, the first four tumors (HNSC, UCEC, LUSC, SKCM) with more than 15% mutation rates were selected based on the survival module of the database. The clinical response of ICIs can be predicted by TMB. The mutations of FAT1 and its function in predicting patient prognosis were analyzed using the 1661 patients/samples of TMB and immunotherapy (MSKCC, Nat Genet 201913) dataset.
TMB and MSI
We retrieved the gene mutation data of 10,114 pan-cancer samples from the TCGA database to calculate the mutation score of each sample and obtained tumor mutation burden. The correlation between the expression of FAT1 and TMB was analyzed using Spearman’s correlation test, and the correlation radar chart was drawn using the fmsb package. The MSI scores of 10,415 tumor samples were downloaded and analyzed, resulting in a radar map of the MSI correlation between FAT1 and tumors.
TME and immune cell infiltration
Based on the TCGA Pan-Cancer (PANCAN, N = 10,535, G = 60,499) dataset in Sangerbox, the samples with zero expression levels were filtered, and the expression values were transformed by log2 (x = 0.001). The stromal, immune, and ESTIMATE scores for each patient in each tumor were calculated using the R software package ESTIMATE14 (https://bioinformatics.mdanderson.org/estimate/), according to gene expression. The immune infiltration scores of 9530 tumor samples from 39 tumor types were obtained. The Spearman’s correlation coefficient of FAT1 expression and TME was calculated based on the psych package’s corr.test function, which helped determine the significant correlation between immune infiltration scores. Similarly, samples with zero expression levels were filtered based on the TCGA TARGET GTEx (PANCAN, N = 19,131, G = 60,499) dataset in Sangerbox, and the expression values were transformed by log2 (x = 0.001). Each patient’s immune cell infiltration score in each tumor was evaluated using the deconvo_CIBERSOR method of the IOBR package15,16, following gene expression. Finally, 22 immune cell infiltration scores were obtained from the 10,063 tumor samples of 44 tumor types. The Spearman’s correlation coefficient of FAT1 expression and immune cell infiltration score in each tumor was also calculated using the psych package’s corr.test function to determine the significantly related immune infiltration score.
Protein expression and protein–protein interaction (PPI)
The Human Protein Atlas (https://www.proteinatlas.org/) database was searched for information about FAT1 protein expression in 17 different types of tumor tissues and paracancerous tissues that had been validated by immunohistochemistry. We also explored the expression of FAT1 in different kinds of single cells. STRING (https://string-db.org/)17 is a known and predicted PPI database that was used to analyze the PPI network of FAT1 in this study. The cytoHubba plug-in of Cytoscape was used to analyze the degree of each protein, and then the PPI network was drawn. We also searched the pathways and genes that interact with FAT1 in UCSC Genome Browser. The Gene Ontology and KEGG Pathway were analyzed in Metascape (https://metascape.org/) based on the interacting proteins in the PPI network.
Protein phosphorylation
PhosphoNET (http://www.phosphonet.ca/) is the world’s largest online repository of general and predictive information about human phosphorylation sites, their evolutionary conservation, the identity of protein kinases that may target these sites, and related phosphate sites. The verified and predicted phosphorylation sites were obtained by searching FAT1. The most significant phosphorylation sites and kinases involved in phosphorylation were predicted by calculating the Kinase Predictor V2 Score by the Kinexus P-Site Prediction algorithm.
Correlation analysis of marker genes
Studies have confirmed that various biological processes are involved in the occurrence and development of malignant tumors. Through the review of the literature, we found that FAT1 can participate in multiple functions. The literature review also helped identify the gene markers in EMT18, tumor hypoxia19, exosome20, tumor immunity21, methylation22,23, and autophagy24,25. The correlation between the mRNA expression levels of FAT1 and these marker genes in pan-cancer was obtained by Spearman correlation analysis with the limma package. The results can predict the potential benefits of FAT1 in these processes and lay a bioinformatics foundation for further studying FAT1 in malignant tumors.
Data analysis
Wilcox and T-tests were used for FAT1 differential expression analysis. Cox proportional hazard regression model and Kaplan–Meier methods were used for survival analysis, and Spearman’s analysis was used for studying correlations. Except for special instructions, P < 0.05 demonstrated that the difference was statistically significant. *P < 0.05. **P < 0.01. ***P < 0.001.
Results
FAT1 expression
According to the subcellular locations from COMPARTMENTS (https://compartments.jensenlab.org/), FAT1 protein is mainly expressed in the nucleus, plasma membrane, and extracellular regions of the cell26,27,28 (Fig. 1A). We found that FAT1 was highly expressed in pan-cancer, except for low expression in some tumors (BRCA, KICH, KIRC, LAML, THCA, UCEC) and no differential expression in some tumors (BLCA, OV, PCPG, SKCM, SARC, TGCT, THYM, UCS) (Fig. 1B–F) based on the computational analysis of FAT1 mRNA expression and multi-database analysis. The analysis of FAT1 expression in different tumor stages revealed its significant difference in various tumors like ACC, COAD, ESCA, KICH, KIRP, LUAD, MESO, SKCM, STAD, and THCA (Fig. 2A). Interestingly, there was no significant difference in FAT1 expression between SKCM tumor and normal tissues. FAT1 protein expression was significantly higher in clear cell renal cell carcinoma, colon cancer, and uterine corpus endometrial carcinoma and was significantly lower in breast cancer. There was no significant difference in FAT1 protein expression in lung adenocarcinoma and ovarian cancer (Fig. 2B). The noteworthy point is that, contrary to our findings, Nantana Kwaepila et al.29 reported that FAT1 in breast cancer immunohistological expression data displayed high expression levels in human tumor samples, possibly due to sample specificity issues. High- or medium-intensity staining (Fig. 2C) was observed in the tumor tissues of glioma, head and neck, thyroid, colorectal, endometrial, liver, urothelial, lung, pancreatic cancers, and lymphomas, indicating that the FAT1 protein was significantly overexpressed in the tumor tissues of these diseases. We also found that FAT1 was highly expressed in many glandular epithelial cells, squamous epithelial cells, and specialized epithelial cells based on the summary of single-cell expression of FAT1. It was also highly expressed in astrocytes, smooth muscle cells, and other cells (Fig. 2D).
Subcellular locations and mRNA expression levels of FAT1. (A) Subcellular locations of FAT1 based on protein expression. (B) mRNA expression levels of FAT1 in pan-cancer. (C, D) mRNA expression levels of FAT1 in 12 tumors. (E) FAT1 expression levels in the dataset of 20 tumors in the Oncomine database. The best gene rank percentile determines the cell color for the analyses within the cell. The red indicates high expression, and the blue indicates low expression. (F) mRNA expression of FAT1 in pan-cancer from the Sangerbox database.
FAT1 multi-omics expression. (A) FAT1 mRNA levels in pan-cancer with differential expression at different stages. (B) Based on CPTAC data, FAT1 protein is expressed at low levels in breast cancer and highly expressed in clear cell renal cell carcinoma, colon cancer, and uterine corpus endometrial carcinoma. (C) Immunohistochemical summary histogram of FAT1 in pan-cancer based on The Human Protein Atlas, different colors represent different tumors. (D) Expression levels of FAT1 in 12 single cell types based on The Human Protein Atlas.
Survival analysis
Combining the OS, DSS, DFI, and PFI results of FAT1 calculated by the Cox proportional hazard regression model (Fig. 3) and Kaplan–Meier (Fig. S1), we found that the high FAT1 mRNA expression predicted a worse prognosis in patients with ACC, BRCA, BLCA, KIRC, SARC, KIRP, MESO, LUSC, LUAD, HNSC, THCA, THYM, PAAD, CESC, OV, SKCM, and UVM. In comparison, it predicted a better prognosis in STAD, STES, ESCA, KIRP, UCS, UCEC, READ, and TGCT.
Mutation analysis
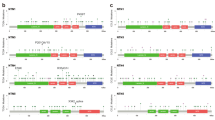
FAT1 encoding gene is located at q35.2 of chromosome 4 (Fig. 4A). We analyzed the mutational information of FAT1 in pan-cancer using cBioportal and found that FAT1 was mutated in more than 10% of 10 tumors, including HNSC and UCEC (Fig. 4B). Missense and truncating were the two most common mutations of FAT1, which primarily affects extracellular and cytoplasmic regions. The PTMs results depicted that FAT1 proteins are primarily phosphorylated and ubiquitinated (Fig. 4C). The mutation survival analysis of the top four tumors with more than 15% mutation rate demonstrated that FAT1 mutations in UCEC predicted better prognosis for tumor patients, but FAT1 mutations in HNSC predicted poor survival (Fig. 4D). The TMB and immunotherapy dataset revealed that FAT1 mutations were observed in 10% of the samples, which is consistent with the findings from the pan-cancer analysis (Fig. 4E). Notably, the OS indicated that patients with FAT1 mutations had a better prognosis (Fig. 4F), suggesting that FAT1 mutations may be used as an indicator of patient’s prognosis.
Mutations of FAT1 in pan-cancer. (A) Location of FAT1 in the chromosome. (B) Mutation of FAT1 in different tumors, “Deep Deletion”, indicates a deep loss, possibly a homozygous deletion, and “Amplification” indicates a high-level amplification (more copies, often focal). (C) General overview of FAT1 mutations in pan-cancer, with nodes and annotations representing different mutation profiles. (D) FAT1 mutation in UCEC predicts a good prognosis for patients, but in HNSC, it predicts a worse prognosis. (E) General overview of FAT mutations in the TMB and immunotherapy (MSKCC, Nat Genet 2019) dataset, with nodes and annotations representing different mutation profiles. (F) FAT1 mutations in the TMB and immunotherapy (MSKCC, Nat Genet 2019) dataset are predicted to have a better prognosis.
Immunocomprehensive analysis
Tumor therapy has advanced significantly owing to immunotherapy that targets immunological checkpoints30. It has been demonstrated that MSI and TMB31 are biomarkers for predicting tumor immunotherapy and are crucial in directing the systemic treatment of tumors32. The analysis of sequencing data demonstrated significant correlations between FAT1 and MSI of several tumors, positive correlations in LUAD, LUSC, and TGCT, and negative correlations in BLCA, DLBC, HNSC, SKCM, and THCA (Fig. 5A). Additionally, FAT1 was also significantly correlated with TMB of many tumors; positively correlated in ACC, KIRC, LAML, PAAD, READ, STAD, and THYM, and negatively correlated in LGG and LUAD (Fig. 5B). The TME has a significant role in tumor occurrence and development, as well as in the evaluation of invasion, metastasis, and prognosis, thus pointing out the future direction for more effective immunotherapy33,34,35. Our study confirmed a significant correlation between the FAT1 expression and TME components in various tumors. Analysis was done to explore the correlation between the FAT1 expression and the Estimate score of the matrix score and immune score. The results revealed that FAT1 was positively correlated with BRCA, DLBC, LGG, OV, and PCPG but negatively correlated with GBM, KIRP, PAAD, PRAD, SARC, SKCM, STES, TGCT, THCA, and THYM (Table 2). Immune cells in the TME are essential for tumor regulation36. This study analyzed the relationship between FAT1 expression and 22 different types of immune cell infiltration in pan-cancer. The results demonstrated that FAT1 was significantly correlated with one or more immune cell infiltration in all tumor types except CHOL. There was a significant positive correlation with T-cells-CD4-memory-resting and macrophages-M1 of THYM and a significant negative correlation with Tregs (Fig. 5C).
PPI and functional enrichment analysis
PPI is significant in a tumor’s molecular biological regulation because of its versatility, specificity, and adaptability17,37. The PPI results based on Cytoscape analysis demonstrated that there was a significant interaction between MYC, EGFR, PIK3CA, TP53, TNF, RELA, NFKBIA, TJP1, JUN, PPARA, and FAT1 (Fig. 6A), and the degree of these interacting proteins in the network was more than 15. Second, PPI results from the UCSC tool depicted that FAT1 interacts with many transcription factors (ADNP, CBX5) and essential proteins such as EIF5B histones, AGL, and HSPA1A (Fig. 6B). The results of Gene Ontology and KEGG Pathway enrichment analysis (https://www.kegg.jp/kegg/kegg1.html)38 based on the proteins in the PPI network revealed that it was significantly enriched in cancer pathways and particularly related to transcription factor binding, intracellular receptor signaling pathway, regulation of epithelial cell proliferation, negative regulation of cell differentiation, epithelial cell development, and other pathways (Fig. 6C,D).
FAT1 protein interaction network and pathway enrichment analysis. (A) Protein–protein interaction network of FAT1 in the STRING database, the darker color of the linkage represents stronger correlation, active interaction sources including text mining, experiments, databases, co-expression, neighborhood, gene fusion, co-occurrence, the minimum required interaction score is 0.4 (medium confidengce). (B) The top 24 genes with the strongest correlation with FAT1 in UCSC, only FAT1-interacting genes and only the most-mentioned/most-curated interactions, Solid grey lines—only text-mining support for this interaction, with the thickness of the line indicating the number of articles supporting it. Dashed blue lines—at least one curated database supports this interaction. Dark blue—the information is derived from a paper describing fewer than ten interactions. Light blue—the information is derived from a high-throughput paper describing more than ten interactions. Solid blue lines—databases and text mining support this interaction and are colored by support. The top 20 results of KEGG and GO enrichment analysis of FAT1 and its interacting proteins, (C) cluster network plot, (D) histogram, different colors represent different pathways and biological processes (The color of panel C corresponds to the color in panel D), “Log10 (P)” is the p-value in log base 10.
FAT1 protein phosphorylation
Protein phosphorylation is recognized as the primary method of protein PTMs, and it is an effective way to regulate protein function under the catalysis of protein kinases. It is a fundamental cause of intracellular processes such as cell growth and development, signal transduction, and metabolism39,40. A database search revealed that 30 FAT1 proteins had been phosphorylated (Table 2). Second, data scoring predicted 45 sites most likely to be phosphorylated, and the protein kinases (Table S1) most likely to participate in the phosphorylation process were also predicted. The phosphorylation of the FAT1 protein is now better understood.
Correlation analysis of tumor marker genes
Groundbreaking research by Ievgenia Pastushenko et al.1 demonstrated that FAT1 mutation promotes tumor initiation, progression, invasion, stemness, and metastasis by inducing hybrid EMT states in mouse and human skin squamous cell carcinoma. Chitrangda Srivastava et al.41 found that FAT1 is a novel regulator of EMT in anoxic GBM, which suggested that it may be a viable therapeutic candidate. According to Xiaoling Hu et al.42, FAT1 destroys the MAPK/ERK pathway and participates in the EMT process of esophageal squamous cell carcinoma. Our analysis of the correlation between FAT1 and recognized EMT transcription factors and receptors demonstrated that FAT1 is associated with EMT marker genes in almost all tumors (Fig. 7), indicating that FAT1 may be involved in the EMT process in all tumors.
Exosomes may affect tumor progression by regulating immune function, promoting tumor angiogenesis and metastasis, and interacting directly with tumor cells43,44,45. It can participate in immune response, antigen presentation, cell migration, cell differentiation, and tumor invasion. Different immune cells are also involved in tumor regulation46. Our results depicted that FAT1 was significantly associated with one or more exosome markers in all tumors (Fig. S2). Immune cell marker genes analysis illustrated that FAT1 was significantly associated with a variety of immune cell markers in BRCA, DLBC, GBM, HNSC, KIRC, KIRP, LIHC, LUAD, LUSC, OV, PCPG, PRAD, SARC, SKCM, STAD, TGCT, THCA, THYM, UCEC, and other tumors.
Both DNA and RNA can be methylated. The family of DNA methyltransferases play a central role in epigenetic regulation23. RNA methylation modification accounts for more than 60% of all RNA modifications, of which m6A is an essential methylation modification in which writers, erasers, and readers play different roles, including regulation of tissue development, circadian rhythm, DNA damage response, sex determination, T cell homeostasis and tumorigenesis22. The correlation between FAT1 expression and methylation marker genes is displayed in Fig. S3. Our results demonstrated a significant correlation between FAT1 and multiple methylation marker genes in different tumors except for CHOL, ESCA, LAML, and UCS.
Hypoxia has recently been confirmed to play an essential role in tumor progression19. Tumor hypoxia is associated with increased invasion and metastasis and shows typical driving mutation characteristics. The correlation between FAT1 expression and tumor hypoxia marker genes is revealed in Fig. S4. The findings revealed a significant correlation between FAT1 and multiple hypoxia-related mutant genes in almost all tumors except for LAML.
The dynamic equilibrium of cells, tissues, and organisms depends heavily on the autophagy pathway, which is mediated by evolutionarily conserved ATGs24. ATGs mutations play an important role in various diseases, including cancer25. The correlation between FAT1 expression and autophagy-related marker genes is illustrated in Fig. S5.
Discussion
FAT atypical cadherin 1 (FAT1) encodes protocadherin, one of the most frequently mutated genes in human cancers. Studies conducted over the past 20 years have demonstrated that FAT1 regulates multiple signaling pathways, including Wnt/β-catenin, Hippo and MAPK/ ERK, to affect the proliferation, migration, and invasion of various tumor cells1,47. In this study, FAT1 was found to have a mutation rate of > 10% in more than ten tumors. It has not been established whether FAT1 plays a role in the pathogenesis of different tumors through some common molecular mechanisms; therefore, we performed a comprehensive pan-cancer analysis of FAT1 based on data in TCGA, CPTAC, and GEO from an overall tumor perspective, focusing on gene expression, gene alterations, protein phosphorylation, and prognosis.
FAT1 is highly expressed in most tumors. However, the prognostic survival analysis of the FAT1 gene data suggested different conclusions for different tumors. In the present study, we combined different prognostic analysis methods and data. Finally, we revealed that high FAT1 expression predicted a worse prognosis in all tumors except STAD, STES, ESCA, KIRP, UCS, UCEC, READ, and TGCT. This is according to certain recent studies47.
According to previous studies, FAT1 is mutated in a variety of tumors. This study analyzed the relationship between FAT1 mutations and patient survival in the four tumors with the highest mutation rates. We revealed that FAT1 mutations in UCEC predicted a better prognosis for tumor patients, whereas they predicted poor survival in HNSC patients. According to the OS, patients with FAT1 mutation had a better prognosis, as revealed by the mutation data from TMB and immunotherapy (MSKCC, NatGenet 2019)13. Furthermore, we found that FAT1 was significantly correlated with TMB in ACC, KIRC, LAML, PAAD, READ, STAD, THYM, LGG, and LUAD tumors. This result suggested that FAT1 mutations may serve as immunotherapeutic markers for these tumors and may be useful for guiding novel immunotherapies.
The PTMs regulate the function of most eukaryotic proteins48, and protein phosphorylation is very closely associated with tumors49. In this study, the database statistically predicted the most phosphorylated sites and protein kinases of FAT1 protein to explore potential associations between FAT1 and tumors. Recent studies have demonstrated that deletion of FAT1 in cutaneous squamous cell carcinoma accelerates tumor initiation, malignant progression and promotes the EMT phenotype1. We found a significant association between FAT1 mRNA level and EMT phenotype-related marker genes in almost all tumor types, implying that FAT1 may play a role in the EMT phenotype in other tumors. Studies analyzing the correlation between FAT1 and markers genes such as exosomal and immune, methylation, hypoxia, and autophagy have produced a number of additional useful findings. These phenomena need to be confirmed by further research.
For the first time, this study provided evidence for a potential correlation between FAT1 expression and MSI or TMB in all TCGA tumors. We also integrated information on FAT1 binding components and FAT1 expression-related genes in all tumors. We performed enrichment analyses to identify the potential effects of FAT1 in tumor pathways, reverse regulation of cell differentiation, transcription factor binding, and epithelial cell development.
FAT1 has been reported to have immunotherapeutic potential in several tumors50,51. We applied the immunological deconvolution method to observe the statistical correlation between FAT1 expression and the infiltration level of 22 immune cells in pan-cancer. FAT1 may play a function in the immunotherapy of THYM as a result of the highly substantial association between THYM and many immune cells in THYM. Additionally, we found that FAT1 expression was significantly and positively correlated with immune scores of BRCA, DLBC, LGG, OV, and PCPG based on the calculated stromal, immune, and ESTIMATE scores for each patient in the TCGA pan-cancer dataset of 9530 tumor samples from a total of 39 tumor types. These findings can provide research directions for tumor immunotherapy based on FAT1.
Our first pan-cancer analysis of FAT1 demonstrated a statistical correlation between FAT1 expression and clinical prognosis, protein phosphorylation, MSI, TMB, TME, and immune cell infiltration, which helped comprehend the role of FAT1 in tumorigenesis from the perspective of clinical tumor samples. Moreover, FAT1 mutations were also found to be closely associated with immunotherapy and may develop into tumor immunotherapy markers. This study also helped to broaden the research on FAT1 in tumors. Undeniably, this study was based on extensive tumor data, but it had the limitation of lacking rigorous experiments to validate the findings. More reliable experiments will be needed to validate the various potential roles of FAT1 in tumors.
Data availability
These data are drawn from the following resources in the public domain. UCSC Xena (http://xena.ucsc.edu/), the cohort is TCGA Pan-Cancer (PANCAN: https://pancanatlas.xenahubs.net). All data in this study were permitted for use.
References
Pastushenko, I. et al. Fat1 deletion promotes hybrid EMT state, tumour stemness and metastasis. Nature 589, 448–455. https://doi.org/10.1038/s41586-020-03046-1 (2021).
Blum, A., Wang, P. & Zenklusen, J. C. SnapShot: TCGA-analyzed tumors. Cell 173, 530. https://doi.org/10.1016/j.cell.2018.03.059 (2018).
Tomczak, K., Czerwińska, P. & Wiznerowicz, M. The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp. Oncol. (Pozn.) 19, A68–A77. https://doi.org/10.5114/wo.2014.47136 (2015).
Barrett, T. et al. NCBI GEO: Archive for functional genomics data sets–update. Nucleic Acids Res. 41, D991–D995. https://doi.org/10.1093/nar/gks1193 (2013).
Campbell, P. J. et al. Pan-cancer analysis of whole genomes. Nature 578, 82–93. https://doi.org/10.1038/s41586-020-1969-6 (2020).
Morris, L. G. T. et al. Recurrent somatic mutation of FAT1 in multiple human cancers leads to aberrant Wnt activation. Nat. Genet. 45, 253–261. https://doi.org/10.1038/ng.2538 (2013).
Tang, Z., Kang, B., Li, C., Chen, T. & Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 47, W556–W560. https://doi.org/10.1093/nar/gkz430 (2019).
Rhodes, D. R. et al. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia 6, 1–6 (2004).
Uhlen, M. et al. A pathology atlas of the human cancer transcriptome. Science https://doi.org/10.1126/science.aan2507 (2017).
Chandrashekar, D. S. et al. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 19, 649–658. https://doi.org/10.1016/j.neo.2017.05.002 (2017).
Liu, J. et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell https://doi.org/10.1016/j.cell.2018.02.052 (2018).
Cerami, E. et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404. https://doi.org/10.1158/2159-8290.CD-12-0095 (2012).
Samstein, R. M. et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 51, 202–206. https://doi.org/10.1038/s41588-018-0312-8 (2019).
Yoshihara, K. et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 4, 2612. https://doi.org/10.1038/ncomms3612 (2013).
Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457. https://doi.org/10.1038/nmeth.3337 (2015).
Zeng, D. et al. IOBR: Multi-omics immuno-oncology biological research to decode tumor microenvironment and signatures. Front. Immunol. 12, 687975. https://doi.org/10.3389/fimmu.2021.687975 (2021).
Szklarczyk, D. et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 49, D605–D612. https://doi.org/10.1093/nar/gkaa1074 (2021).
Lamouille, S., Xu, J. & Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15, 178–196. https://doi.org/10.1038/nrm3758 (2014).
Bhandari, V. et al. Molecular landmarks of tumor hypoxia across cancer types. Nat. Genet. 51, 308–318. https://doi.org/10.1038/s41588-018-0318-2 (2019).
Deng, F. & Miller, J. A review on protein markers of exosome from different bio-resources and the antibodies used for characterization. J. Histotechnol. 42, 226–239. https://doi.org/10.1080/01478885.2019.1646984 (2019).
Burugu, S., Dancsok, A. R. & Nielsen, T. O. Emerging targets in cancer immunotherapy. Semin. Cancer Biol. 52, 39–52. https://doi.org/10.1016/j.semcancer.2017.10.001 (2018).
Liu, J., Harada, B. T. & He, C. Regulation of gene expression by N-methyladenosine in cancer. Trends Cell Biol. 29, 487–499. https://doi.org/10.1016/j.tcb.2019.02.008 (2019).
Lyko, F. The DNA methyltransferase family: A versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 19, 81–92. https://doi.org/10.1038/nrg.2017.80 (2018).
Galluzzi, L. et al. Molecular definitions of autophagy and related processes. EMBO J 36, 1811–1836. https://doi.org/10.15252/embj.201796697 (2017).
Levine, B. & Kroemer, G. Biological functions of autophagy genes: A disease perspective. Cell 176, 11–42. https://doi.org/10.1016/j.cell.2018.09.048 (2019).
Dunne, J. et al. Molecular cloning and tissue expression of FAT, the human homologue of the Drosophila fat gene that is located on chromosome 4q34-q35 and encodes a putative adhesion molecule. Genomics 30, 207–223. https://doi.org/10.1006/geno.1995.9884 (1995).
Magg, T., Schreiner, D., Solis, G. P., Bade, E. G. & Hofer, H. W. Processing of the human protocadherin Fat1 and translocation of its cytoplasmic domain to the nucleus. Exp. Cell Res. 307, 100–108. https://doi.org/10.1016/j.yexcr.2005.03.006 (2005).
Tanoue, T. & Takeichi, M. Mammalian Fat1 cadherin regulates actin dynamics and cell-cell contact. J. Cell Biol. 165, 517–528. https://doi.org/10.1083/jcb.200403006 (2004).
Kwaepila, N., Burns, G. & Leong, A. S. Immunohistological localisation of human FAT1 (hFAT) protein in 326 breast cancers. Does this adhesion molecule have a role in pathogenesis?. Pathology 38, 125–131. https://doi.org/10.1080/00313020600559975 (2006).
Carlino, M. S., Larkin, J. & Long, G. V. Immune checkpoint inhibitors in melanoma. Lancet 398, 1002–1014. https://doi.org/10.1016/S0140-6736(21)01206-X (2021).
Rizvi, N. A. et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128. https://doi.org/10.1126/science.aaa1348 (2015).
Joshi, S. S. & Badgwell, B. D. Current treatment and recent progress in gastric cancer. CA Cancer J. Clin. 71, 264–279. https://doi.org/10.3322/caac.21657 (2021).
Barker, H. E., Paget, J. T. E., Khan, A. A. & Harrington, K. J. The tumour microenvironment after radiotherapy: Mechanisms of resistance and recurrence. Nat. Rev. Cancer 15, 409–425. https://doi.org/10.1038/nrc3958 (2015).
Kubli, S. P., Berger, T., Araujo, D. V., Siu, L. L. & Mak, T. W. Beyond immune checkpoint blockade: Emerging immunological strategies. Nat. Rev. Drug Discov. 20, 899–919. https://doi.org/10.1038/s41573-021-00155-y (2021).
Maman, S. & Witz, I. P. A history of exploring cancer in context. Nat. Rev. Cancer 18, 359–376. https://doi.org/10.1038/s41568-018-0006-7 (2018).
Hanahan, D. & Coussens, L. M. Accessories to the crime: Functions of cells recruited to the tumor microenvironment. Cancer Cell 21, 309–322. https://doi.org/10.1016/j.ccr.2012.02.022 (2012).
Cheng, F. et al. Comprehensive characterization of protein-protein interactions perturbed by disease mutations. Nat. Genet. 53, 342–353. https://doi.org/10.1038/s41588-020-00774-y (2021).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Cohen, P. Protein kinases–the major drug targets of the twenty-first century?. Nat. Rev Drug. Discov. 1, 309–315 (2002).
Ubersax, J. A. & Ferrell, J. E. Mechanisms of specificity in protein phosphorylation. Nat. Rev. Mol. Cell Biol. 8, 530–541 (2007).
Srivastava, C. et al. FAT1 modulates EMT and stemness genes expression in hypoxic glioblastoma. Int. J. Cancer 142, 805–812. https://doi.org/10.1002/ijc.31092 (2018).
Hu, X. et al. FAT1 prevents epithelial mesenchymal transition (EMT) via MAPK/ERK signaling pathway in esophageal squamous cell cancer. Cancer Lett. 397, 83–93. https://doi.org/10.1016/j.canlet.2017.03.033 (2017).
Liu, J. et al. The biology, function, and applications of exosomes in cancer. Acta Pharm. Sin. B 11, 2783–2797. https://doi.org/10.1016/j.apsb.2021.01.001 (2021).
Théry, C., Zitvogel, L. & Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2, 569–579. https://doi.org/10.1038/nri855 (2002).
Xia, H., Green, D. R. & Zou, W. Autophagy in tumour immunity and therapy. Nat. Rev. Cancer 21, 281–297. https://doi.org/10.1038/s41568-021-00344-2 (2021).
Elinav, E. et al. Inflammation-induced cancer: Crosstalk between tumours, immune cells and microorganisms. Nat. Rev. Cancer 13, 759–771. https://doi.org/10.1038/nrc3611 (2013).
Peng, Z., Gong, Y. & Liang, X. Role of FAT1 in health and disease. Oncol. Lett. 21, 398. https://doi.org/10.3892/ol.2021.12659 (2021).
Mann, M. & Jensen, O. N. Proteomic analysis of post-translational modifications. Nat. Biotechnol. 21, 255–261 (2003).
Singh, V. et al. Phosphorylation: Implications in cancer. Protein J. 36, 1–6. https://doi.org/10.1007/s10930-017-9696-z (2017).
Chaudhary, S. et al. Differential mutation spectrum and immune landscape in African Americans versus Whites: A possible determinant to health disparity in head and neck cancer. Cancer Lett. 492, 44–53. https://doi.org/10.1016/j.canlet.2020.07.029 (2020).
Grandi, A. et al. Vaccination with a FAT1-derived B cell epitope combined with tumor-specific B and T cell epitopes elicits additive protection in cancer mouse models. Front. Oncol. 8, 481. https://doi.org/10.3389/fonc.2018.00481 (2018).
Acknowledgements
This work was funded by the Special Foundation for Postgraduate Innovation of Jiangxi Province (grant no. YC2021-S791).
Author information
Authors and Affiliations
Contributions
Z.W. and K.L. performed the research. K.L. and H.X. designed the research study. Z.W. analyzed the data. Z.W. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, Z., Lin, K. & Xiao, H. A pan-cancer analysis of the FAT1 in human tumors. Sci Rep 12, 21598 (2022). https://doi.org/10.1038/s41598-022-26008-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-26008-1
- Springer Nature Limited