Abstract
The broadly neutralizing antibody (bNAb) CAP256-VRC26.25 has exceptional potency against HIV-1 and has been considered for clinical use. During the characterization and production of this bNAb, we observed several unusual features. First, the antibody appeared to adhere to pipette tips, requiring tips to be changed during serial dilution to accurately measure potency. Second, during production scale-up, proteolytic cleavage was discovered to target an extended heavy chain loop, which was attributed to a protease in spent medium from 2-week culture. To enable large scale production, we altered the site of cleavage via a single amino acid change, K100mA. The resultant antibody retained potency and breadth while avoiding protease cleavage. We also added the half-life extending mutation LS, which improved the in vivo persistence in animal models, but did not impact neutralization activity; we observed the same preservation of neutralization for bNAbs VRC01, N6, and PGDM1400 with LS on a 208-virus panel. The final engineered antibody, CAP256V2LS, retained the extraordinary neutralization potency of the parental antibody, had a favorable pharmacokinetic profile in animal models, and was negative in in vitro assessment of autoreactivity. CAP256V2LS has the requisite potency, developability and suitability for scale-up, allowing its advancement as a clinical candidate.
Similar content being viewed by others
Introduction
Broadly neutralizing antibodies (bNAbs) against HIV are potentially useful for prevention or treatment of infection1. The recent AMP clinical trial tested the effect of the bNAb VRC01 for prevention of infection; while there was no overall efficacy, VRC01 blocked infection of the most antibody-sensitive viruses, with 75% efficacy against viruses that are neutralized with an in vitro ID80 of less than 1 μg/ml2. This result suggests that bNAbs with greater breadth and potency than VRC01 might be efficacious for HIV prevention. In addition, to maximize the coverage of diverse HIV strains, and minimize the effect of escape mutations at single epitopes, it is likely that multiple bNAbs targeting different epitopes would need to be used in combination. CAP256-VRC26.25 is an exceptionally potent bNAb, and is particularly broad and potent against non-B clade virus strains3. A study of bNAbs assessed against viruses of clade C, the most common clade worldwide, found that the most favorable combinations of bNAbs included CAP256-VRC26.254. Therefore, production of CAP256-VRC26.25 has been a priority.
While CAP256-VRC26.25 can be easily produced in HEK-Expi293F cell cultures grown for up to 6 days, as is commonly done in research laboratories, large-scale production of CAP256-VRC26.25 was hampered by proteolytic cleavage, also referred to as clipping, of the antibody when produced in CHO cell culture for 14–17 days. This cleavage was attributed to a residual protease present in the spent culture medium, which attacks an exposed lysine residue in the highly extended CDRH3 loop of the heavy chain5. Multiple process strategies were evaluated to minimize the proteolytic cleavage observed during cell culture6,7. These included (1) a fill-draw approach that resembled a repeated fed batch, where 75% of the bioreactor was harvested on day 10 and refilled with fresh media every 3 days (2) inclusion of protease inhibitors for serine proteases8,9 in the cell culture to effectively inhibit proteolytic activity and (3) alternating tangential flow (ATF)-Perfusion cell culture10,11, allowing continuous antibody harvest. All three approaches minimized CAP256 cleavage below 5%; however, due to risks associated with scaling and transferring these processes, product stability risks, and long-term development risks, it was determined that engineering out the proteolytic cleavage site was better suited for long term clinical product development.
Here, we show that a single amino acid change in the CDRH3 was sufficient to prevent cleavage of CAP256-VRC26.25 while retaining neutralization activity. Further addition of the half-life extending mutation, LS, improved the pharmacokinetic profile in model animals with no impact on neutralization. The resulting modified antibody, CAP256V2LS, was suitable for scale-up to 2000 liter cultures, allowing its advancement to a promising clinical candidate.
Results
Mutant screening
Previous work identified a site of proteolytic cleavage (clipping) at the lysine at position K100m (Kabat numbering) in the CAP256-VRC26.25 CDRH35 (Fig. 1a,b). We therefore generated a series of mutants in CAP256-VRC26.25 heavy chain in which that position was changed to every possible residue except cysteine. The antibodies were tested against a panel of Env-pseudoviruses to check for loss of neutralization (Fig. 1c, Suppl Fig. S1A). Neutralization of Env-pseudovirus CAP256.209.c9 was unaffected by any of the mutations. Activity of most mutants against Env-pseudoviruses BG505 and DU156 was decreased five to 100-fold; the least affected mutants were further tested against MB539, TH976, PVO.04, and AC10.29. Mutation to alanine, methionine, or glutamine (K100mA, K100mM, and K100mQ) caused a small (< fivefold) overall reduction in potency, while the other mutations greatly reduced or abrogated activity. Changing the lysine to an arginine (K100mR) did not alter the neutralization activity. Of note, the CAP256-VRC26.25 appeared to be carried over in pipet tips during dilution, causing an artifact in which the neutralization activity does not titer out. This led us to adopt a neutralization assay protocol in which pipet tips are changed after each antibody dilution step; doing so was found to be critical for accurate assessment of CAP256-VRC26.25 neutralization potency (Suppl Fig. S2).
CAP256-VRC26.25 mutations at the proteolytic cleavage site have a variable effect on neutralization potency. (a) CDRH3 sequence of wild-type CAP256-VRC26.25. The arrow and slash indicate the site of cleavage. The positions are labeled according to the Kabat numbering scheme. Cysteines involved in a disulfide bond are highlighted in green. (b) The wild-type CAP256-VRC26.25 Fab is shown (PDB ID 5DT1) with the CDRH3 highlighted to show the location of the cleavage between K100m and Q100n and its relation to the disulfide bond (green) and sulfated tyrosines (stick representation). (c) Neutralization of Env-pseudovirus panel by CAP256.25 variants. Each Ab was expressed using the indicated CAP256-VRC26.25 heavy chain paired with wild type light chain. nd not done. Data are IC50 in μg/ml.
To mimic the exposure to proteases released by dead cells in 19-days CHO cell culture, we passed medium from 19 days CAP256-VRC26.25 CHO cell cultures over protein A and retained the flow-through fraction. The wild-type antibody and mutants K100mA, K100mM, K100mQ, and K100mR (produced in 6 days HEK-Expi293F cultures) were incubated in this protease-containing flow-through material at 37 °C for 0 or 72 h. After additional protein A column purification, treated or untreated mAbs were tested by reducing CGE-SDS (GX II) chromatography analysis to monitor heavy chain cleavage12. The chromatography trace for wild-type CAP256-VRC26.25 included extra peaks identified as the products of heavy-chain cleavage; 6.4% of the antibody mass was assigned to the clipped products (Fig. 2). We observed that the K100mR mutant was also subject to cleavage; this was not unexpected, as lysine and arginine are both targets of serine proteases, the presumptive class of proteases acting on the antibodies. In contrast, the K100mA, K100mM, and K100mQ variants were not cleaved (Fig. 2).
CAP256-VRC26.25 variants resist cleavage. (a) Samples were treated with day 19 CHO supernatant flow-through containing protease (Day 19FT) for 0 or 72 h. Treated and untreated samples were separated by size using reducing capillary gel electrophoresis. Peaks labelled HCC1, HCC2 contain clipped product; HC contains unclipped heavy chain, LC contains light chain. (b) % Cleaved heavy chain showing 0 and 72 h treatments. % HCC was calculated as area of each peak divided by total area of all peaks.
The cleavage–resistant variant that showed the greatest potency in the initial neutralization screen, K100mA, was further tested on a 208 multiclade Env-pseudovirus panel. When compared to wild type, this mutant had the same overall breadth and median potency (Fig. 3a, Table 1, Fig. S1, Table S1). At the level of individual viruses, we observed that K100mA gained potency for some viruses and lost potency for others, but in a balanced manner such that the total fraction of viruses sensitive to K100mA was the same as to wild type (Fig. S1B). We noted a very high correlation between IC50 values for the two antibodies (Spearman’s rho 0.96, p < 0.0001) as well as for IC80 values (Spearman’s rho 0.97, p < 0.0001). In addition, we used the GPS-TSP algorithm13 to predict the impact of K100mA on tyrosine sulfation of the CDRH3, as this post-translational modification is critical for full potency of the antibody (Cai et al., manuscript submitted). Predicted sulfation of wild-type and K100mA were nearly identical (Fig. S1C), in agreement with the observed retention of neutralization activity. Because the K100mA mutant retained the full breadth and potency of the wild type, while resisting proteolytic cleavage, we selected this variant for further development.
K100mA mutant maintains breadth and potency as assessed on large virus panels. (a) Neutralization by CAP256-VRC26.25 wild type, K100mA, and CAP256V2LS was measured against a multi-clade 208 Env-pseudovirus panel (left) and 100 acute Clade C Env-pseudoviruses (right). Each dot represents one pseudovirus. (b) Dendrogram shows phylogenetic relatedness of Env sequences in multi-clade panel, color-coded by neutralization sensitivity.
Half-life extending mutation in lead variant
In addition to the K100mA mutation, we added the LS mutation in the Fc portion of the heavy chain, which was previously shown to increase the half-life of parental CAP256-VRC26.25 upon passive administration in non-human primates14. In humans, the LS mutations in other antibodies have been shown to increase serum half-life in vivo; the mechanism is related to increased antibody binding to the FcRn receptor15,16. LS is not expected to affect neutralization potency. To verify this, we assessed neutralization on the 208 multiclade Env-pseudovirus panel for three pairs of bNAbs: VRC01 and VRC01LS, N6 and N6-LS, and PGDM1400 and PGDM1400LS (Suppl Table S1E). As expected, only very minor differences were noted between the antibodies with and without LS (Table 1, Suppl Table S1A,B), with highly significant correlation between values: for each pair of antibodies, both IC50 values and IC80 values correlated with Spearman’s rho of 0.94 or higher and p < 0.0001; and the median IC50s were no more than twofold different, which is within experimental error for this assay17. We therefore proceeded to add LS to the Fc portion of the CAP256-VRC26.25.K100mA construct.
The resulting construct, CAP256V2LS, was produced at high purity under GMP conditions. This clinical-grade product was assessed for neutralization on the 208 multiclade Env-pseudovirus panel and was slightly more potent and broad than research lots of either wild type or K100mA (Fig. 3, Suppl Fig. S3, Table 1, Suppl Table S1A,B). This improvement may be due to enhanced purity of the clinical-grade product compared to research laboratory stocks, and/or improved tyrosine sulfation in the CHO cell line used for production18. Overall, clinical-grade CAP256V2LS is 63% broad on 208 viruses, with high potency and 70% breadth across non-B clade viruses, at a cutoff of IC50 < 50 μg/ml. At the more stringent cutoff of ID80 < 1 μg/ml suggested by the AMP trial results, breadth on non-clade B was 44% overall and 59% against clade C pseudoviruses in the panel (Fig. 3, Table S1A,B). The potency of CAP256V2LS is greater than other HIV-1 bNAbs that have been in clinical trials (Table S1A,B); and its breadth, while lower than some of them, is similar to mAb 10–1074. GMP-grade CAP256V2LS was also tested on a panel of 100 clade C viruses, representing the dominant sequences in southern Africa, with 62% breadth at ID80 < 1 and a median IC50 of 0.002 and ID80 of 0.007 μg/ml (Fig. 3, Suppl Table S1C,D). The combination with VRC07-523-LS had a predicted breadth of 90% at ID80 < 1 on both the multiclade and the clade C panels (Table 1, Suppl Table S1B,D).
Structural analysis of mutant
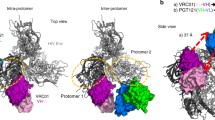
To understand why the K100mA mutation had only minor effects on neutralization breadth and potency, we examined the structure of wild-type CAP256-VRC26.25 alone and in complex with a closed prefusion Env trimer (Fig. 4a,b). In the unliganded Fab structure3, K100m formed an electrostatic interaction with D100g of the CDRH3, while in the bound structure19 the D100g residue of the CDRH3 interacted with the positively charged K169 residue of the HIV-1 Env, thereby breaking contact with K100m (Fig. 4b). The location and orientation of the wild-type K100m outside of the apex cavity and facing the solvent prevents any meaningful contact with the Env trimer beyond minimal buried surface area of under 10 Å2 on the backbone. The side chain faced away from the Env apex cavity and away from any potential Env interactions. As expected from this observation, modelling of Ala at position 100 m onto the cryo-EM structure resulted in no change in the Env binding interactions (Fig. 4c,d).
Position 100m in the CAP256-VRC26.25 CDRH3 does not contact the HIV-1 Env. (a) Cryo-EM structure of CAP256-VRC26.25 in complex with the CAP256.wk34.c80 SOSIP.RnS2 Env trimer. Trimer is shown in surface rendering, with protomers in grey, pink, and light blue; glycans in green; antibody heavy chain in yellow ribbon, light chain in slate ribbon. (b) Close-up of the unliganded CDRH3 conformation, highlighting the K100m contact with D100g (top) in contrast to the bound Fab where D100g is making electrostatic contacts with K169 of the Env. (c) (left) K100m is located above the central cavity in the Env apex facing the solvent. (right) modeled alanine at position 100m indicates no addition or loss of Env contacts. (d) The central cavity relative to position 100m is shown at a 90° rotation from (c).
Pharmacokinetic and autoreactivity analyses
Since CAP256V2LS is of interest for clinical use in HIV prevention, we tested its pharmacokinetic (pK) profile in rhesus macaques and in FcRn mice. The latter are transgenic mice expressing the human FcRn receptor and are commonly used to assess serum half-life of monoclonal antibodies20. Rhesus macaques were infused with 10 mg/kg of CAP256V2LS and parental CAP256-VRC26.25 with LS (CAP256LS); antibody concentrations above 10 μg/ml were maintained for three weeks post-infusion for both constructs (Fig. 5a). This is similar to published values for VRC07-523LS, which is a potent CD4 binding site antibody that is under clinical investigation for both HIV-1 prevention and therapy21. Concordant data were observed in a second model, human FcRn transgenic mice, in which CAP256V2LS had pK curves similar to VRC07-523LS (Fig. 5b). We also tested for autoreactivity in vitro, which can be predictive of poor pharmacokinetic properties in vivo22. CAP256V2LS was negative in cardiolipin binding and Hep-2 cell staining, indicating a lack of autoreactivity (Fig. 5c,d). We did not observe any anti-drug activity (ADA) in the rhesus pK studies.
CAP256V2LS has a favorable pharmacokinetic (PK) profile in animal models and no autoreactivity in vitro. (a) PK in rhesus macaques (non-human primates, NHP). Naïve rhesus macaques were infused with 10 mg/kg of antibody, and the antibody concentrations were measured by an anti-idiotype based ELISA. Each line represents one animal. (b) PK in human FcRn mice. Naïve human FcRn mice were infused with 5 mg/kg of CAP256V2LS or VRC07-523LS and antibody concentrations were measured by an anti-idiotype based ELISA. (c) Hep-2 cell staining. (d) Cardiolipin binding at 100 and 33 μg/ml inputs; values shown as GPL units. VRC01-LS, 4E10, VRC07-523-LS, and VRC07-G54W are included as controls in (c,d).
Discussion
Proteolytic cleavage during large-scale production of antibodies can cause severe loss of product and reduced potency, a major roadblock to clinical advancement23. We found a single mutation in CAP256-VRC26.25 that conferred both resistance to cleavage and retention of neutralization activity. The K100mA mutation allows production at large scale of this highly potent bNAb. This mutation was paired with the LS mutation in Fc, and the resulting CAP256V2LS antibody has a pharmacokinetic profile in NHP and FcRn mice similar to VRC07-523LS, which has a half-life of about 40 days in humans24. CAP256V2LS was negative in tests for autoreactivity, also a favorable finding for clinical use22.
Modelling of the K100mA mutation onto the cryo-EM structure suggests that the amino acid at this position has minimal contacts with the Env trimer, explaining the retention of full activity. It is also possible that there are transient interactions with the mobile N160 glycans in the region, although no contacts were observed in the structure with the native K100m. Despite the lack of contact with trimer at this position, certain amino acids did reduce neutralization activity. Indirect structural effects may explain two of the mutations that had the greatest impact on neutralization: glycine and proline at K100m may affect the peptide backbone flexibility, and thus may reduce the ability of the CDRH3 loop to insert into the trimer apex cavity.
The discovery of a single mutation that mitigates the proteolytic cleavage while retaining neutralization breadth and potency has allowed large-scale GMP production of the clinical candidate CAP256V2LS to proceed. This antibody is undergoing clinical testing for safety and pharmacokinetics in humans25, alone and in combination with VRC07-523-LS (Table S1). CAP256V2LS has the potential to be a valuable weapon in the fight against the HIV-1 pandemic.
Methods
Cleavage monitoring and quantification
To model the conditions of proteolytic cleavage in 19 days cell culture, we first generated protease-containing medium as follows: CHO cells transfected with CAP256V2LS plasmids were grown for 19 days in a Fed-batch bioreactor and harvested on 19th day. The harvest was subjected to protein A column to remove the antibody, and flow through cell culture media was collected and saved as day 19 cell culture harvest supernatant (Day19FT), which contains active protease. To assess the level of cleavage, 0.5 ml of 2 mg/ml antibody sample was incubated with 1.5 ml of Day19FT at 37 °C for 72 h, or immediately chilled at 4 °C (0 h) as a control; and then purified using small ProA columns. The purified mAbs were then tested on GX II under reducing conditions to monitor % heavy chain cleavage12. For GXII assay, 8ug of protein was treated with 30 mM DTT (reducing) in sample buffer containing SDS and were denatured and reduced at 90 °C for 5 min. Protein was then subjected to capillary gel electrophoresis (CGE-SDS) and results were analyzed using Perkin Elmer GXII software. %Heavy chain cleaved peaks were calculated by relative peak area of cleaved peaks to total peak area of heavy and light chain in the reduced sample.
Mutagenesis and protein production
Antibody heavy chain and light chain expression constructs were generated through site-directed mutagenesis (GeneImmune Biotechnology). The antibody variants were expressed by transient transfection in HEK-Expi293F cells (Thermo Fisher) using Turbo293 transfection reagent (SPEED BioSystems) according to the manufacturer’s recommendation. 25 µg plasmid encoding heavy-chain and 25 µg plasmid encoding light-chain variant genes were mixed with 150 ml transfection reagents, added to 50 ml of Expi293 cells at 2.5 × 106 per microliter, and incubated in a shaker incubator at 120 rpm, 37 °C, 9% CO2. At 5 days post-transfection, cell culture supernatant was harvested and purified with a Protein A (GE Healthcare) column. The antibodies were eluted using IgG Elution Buffer (Thermo Fisher) and were brought to neutral pH with 1 M Tris–HCl, pH 8.0. Eluted antibodies were dialyzed against PBS overnight and were evaluated by SDS-PAGE before use.
50 liter production to clinical GMP production
The CAP256V2LS fed-batch process was developed at the 50 L scale and successfully scaled-up to a 2000 L bioreactor for GMP production of clinical trial material using chemically defined, animal component-free medium. Briefly, one vial of the master cell bank is thawed in shake flasks, expanded to a 250 L Single-use Bioreactor (SUB) prior to inoculation of the 2000 L production bioreactor. Purification included a combination of depth filtration, three chromatography steps, viral inactivation and filtration, diafiltration, formulation and concentration. Analytical testing assured product quality. CAP256V2LS was produced from a single cell clone that ranged in productivity from 2.4 to 2.9 g/L titers in clinical material production runs.
mAb neutralization
Neutralization was assessed in one of three formats of the Env-pseudotyped assay17, all of which yield highly similar results.
-
(1)
Screening of variants was performed in 96-well format as follows: 10 μl of fivefold serially diluted mAbs in cDMEM was incubated with 40 μl of diluted HIV-1 Env-pseudotyped virus and incubated for 30 min at 37 °C in a 96-well CulturPlate (Perkin Elmer). 20 μl of TZM-bl cells (10,000 cells/well) with or without 70 μg/ml DEAE-Dextran was then added and incubated overnight at 37 °C. Each experiment plate also had a column of cells only (no Ab or virus) and a column of virus only (no Ab) as controls for background TZM-bl luciferase activity and maximal viral entry, respectively. Serial dilutions were performed with a change of tips at each dilution step to prevent carryover; the importance of this change is illustrated in Fig. S2. The following day, all wells received 100 μl of fresh cDMEM and were incubated overnight at 37 °C. The following day, 50 μl of Steadylite Plus Reporter Gene Assay System (PerkinElmer) was added to all wells, and plates were shaken at 600RPM for 15 min. Luminometry was then performed on a SpectraMax L (Molecular Devices) luminometer. Percent neutralization is determined by calculating the difference in average Relative Light Units (RLU) between virus only wells (cells + virus column) and test wells (cells + plasma/Ab sample + virus), dividing this result by the average RLU of virus only wells (cell + virus column) and multiplying by 100. Background is subtracted from all test wells using the average RLU from the uninfected control wells (cells only column) before calculating the percent neutralization. Neutralizing plasma antibody titers are expressed as the antibody concentration required to achieve 50% neutralization and calculated using a dose–response curve fit with a 5-parameter nonlinear function.
-
(2)
Select monoclonal antibodies were assessed on a panel of 208 geographically and genetically diverse Env pseudoviruses representing the major subtypes and circulating recombinant forms26. Assays were performed by microneutralization in an optimized and qualified automated 384-well format as described17, with the addition of changing tips after each antibody dilution. Data were analyzed as above.
-
(3)
CAP256V2LS was assessed on a panel of 100 clade C Env pseudoviruses from acute infection4. Neutralization assays were conducted using TZM.bl cells as previously described17. Briefly, mAb samples were tested in duplicate in 96-well plates using a primary concentration of 50 μg/ml or 1 μg/ml and serially diluted fivefold seven times. The 1 μg/ml start value was used instead of changing tips, as noted above. HIV-1 Env pseudovirus was added to antibody serial dilutions and plates were incubated for 1 h at 37 °C. TZM.bl cells were then added at 1 × 104/well with DEAE-Dextran at a final concentration of 11 μg/ml. After 48 h incubation at 37 °C, plates were harvested using Promega Bright-Glo luciferase (Madison, WI) and luminescence detected using a Promega GloMax Navigator luminometer. Antibody concentrations that inhibited 50% or 80% of viral infection were determined (IC50 and IC80 titers, respectively). Neutralization assays were conducted in a laboratory meeting Good Clinical Laboratory Practice quality assurance criteria.
Modeling
PDB ID 5DT1 was used for the unliganded CAP256-VRC26.25 Fab and PDB ID 6VTT was used for the CAP256-VRC26.25 Env complex structure. Mutations were made using Coot v0.9 and figures made using Pymol v2.4.2. Refinement, simulated annealing, and MDFF were performed with the mutated coordinates with density from the WT structure using Phenix v1.19 and Isolde v1.1 in ChimeraX v1.1.1 with no significant change in the position of A100m compared to the WT K100m. The trimer in the complex, CAP256.wk34.c80 SOSIP.RnS2, has a sequence derived from an Env clone from the CAP256 donor19 likely to have initiated the CAP256-VRC26 lineage27.
Tyrosine sulfation prediction
Tyrosine sulfation within the CDRH3 was predicted from the amino acid sequence using GPS-TSP software available at http://tsp.biocuckoo.org/13.
Autoreactivity
Autoreactivity was determined by ANA Hep-2 Staining Analysis (ZEUS Scientific Cat. No: FA2400) and anticardiolipin ELISA (Inova Diagnostics Cat. No.: 708625). For the Hep-2 assay, all antibodies were tested at 25 and 50 μg/ml as per manufacturer’s protocol and imaged on a Nikon Ts2R microscope for 500 ms. Scores from 0 to 3 were defined with four control antibodies VRC01-LS, 4E10, VRC07-523LS, and VRC07-G54W. Test antibodies were scored by visual estimation of staining intensity in comparison to the control antibodies. Scores equal to or greater than 1 at 25 μg/ml were classified as autoreactive, and between 0 and 1 as mildly autoreactive. In the cardiolipin ELISA, antibodies were tested at a starting concentration of 100 μg/ml, followed by threefold dilutions. IgG phospholipid (GPL) units were calculated from the standard curve. GPL score < 20 was considered as not reactive, 20–80 as low positive and > 80 as high positive.
Pharmacokinetic studies in NHPs and human FcRn mice
Eleven rhesus macaques of Indian origin (Macaca mulatta) comprising of seven male and four female animals with mean age of 7.2 ± 5.2 years and mean weight of 6.5 ± 3.3 kg were used in this study. Out of these animals, four male animals were infused with VRC07-523LS; two male and two female animals with CAP256LS; and one male and two female animals with CAP256V2LS. All infusions were done intravenously with a 10 mg/kg dose for each mAb. Endotoxin levels were measured for each antibody preparation by the QCL-1000™ endpoint chromogenic LAL assay (Lonza) and were all below 0.5 EU/mg levels. Serum samples were collected prior to injection, and at time points 0, 30 min, 6 h, 12 h, and days 1, 2, 4, 7, 14, 21, 28 and 35. All the sampling, weighing and infusion procedures were done on anaesthetized animals using either ketamine or ketamine/dexmedetomidine cocktail as the anaesthesia agent. Serum was separated by centrifugation. Serum samples were heat inactivated at 56 °C for 30 min, and lipoproteins were pelleted.
Serum mAb levels were determined by an anti-idiotype based ELISA as described previously21. All animals were housed and cared for in accordance with Guide for Care and Use of Laboratory Animals Report number NIH 82-53 (Department of Health and Human Services, Bethesda, Maryland, USA, 1985) in a biosafety level 2 National Institute of Allergy and Infectious Diseases (NIAID) facility. All animal procedures and experiments were performed according to protocols approved by the Institutional Animal Care and Use Committee of the National Institute of Allergy and Infectious Diseases (NIH).
Male human FcRn transgenic mice (C57BL/6, B6.mFcRn−/−hFCRN Tg32 line from The Jackson Laboratory) with mean age of 10.9 ± 1.2 weeks and mean weight of 22.2 ± 3.2 g were used to assess the pharmacokinetics of CAP256V2LS and VRC07-523LS antibodies. Each animal was infused intravenously with 5 mg mAb/kg of body weight. Whole blood samples were collected at day 1, 2, 5, 7, 9, 14, 21, 28 and 35. Serum was separated by centrifugation. Serum mAb levels were measured by an anti-idiotype based ELISA as described previously21. All mice were bred and maintained under pathogen-free conditions at an American Association for the Accreditation of Laboratory Animal Care (AAALAC)-accredited animal facility at the NIAID and housed in accordance with the procedures outlined in the Guide for the Care and Use of Laboratory Animals. All mice were between 6 and 13 weeks of age. The study protocol was evaluated and approved by the NIH Animal Care and Use Committee (ASP VRC-18-747). This study is reported in accordance with ARRIVE guidelines.
Data availability
Neutralization data as reported in Table S1A–D are also available through the CATNAP database, https://www.hiv.lanl.gov/components/sequence/HIV/neutralization/.
References
Miner, M. D., Corey, L. & Montefiori, D. Broadly neutralizing monoclonal antibodies for HIV prevention. J. Int. AIDS Soc. 24(Suppl 7), e25829 (2021).
Corey, L. et al. Two randomized trials of neutralizing antibodies to prevent HIV-1 acquisition. N. Engl. J. Med. 384, 1003–1014 (2021).
Doria-Rose, N. A. et al. New member of the V1V2-directed CAP256-VRC26 lineage that shows increased breadth and exceptional potency. J. Virol. 90, 76–91 (2016).
Wagh, K. et al. Optimal combinations of broadly neutralizing antibodies for prevention and treatment of HIV-1 clade C infection. PLoS Pathog. 12, e1005520 (2016).
Ivleva, V. B. et al. Investigation of sequence clipping and structural heterogeneity of an HIV broadly neutralizing antibody by a comprehensive LC-MS analysis. J. Am. Soc. Mass Spectrom. 29, 1512–1523 (2018).
Fan, Y., Ley, D. & Andersen, M. R. Fed-batch CHO cell culture for lab-scale antibody production. Methods Mol. Biol. 1674, 147–161 (2018).
Martinez-Monge, I. et al. A new strategy for fed-batch process control of HEK293 cell cultures based on alkali buffer addition monitoring: Comparison with O.U.R. dynamic method. Appl. Microbiol. Biotechnol. 102, 10469–10483 (2018).
Cai, C. X. et al. Characterization of AEBSF-antibody modifications for a protease inhibitor supplementation strategy. Anal. Bioanal. Chem. 411, 6111–6118 (2019).
Sreelatha, L. et al. Serine protease inhibitor AEBSF reduces dengue virus infection via decreased cholesterol synthesis. Virus Res. 271, 197672 (2019).
Kelly, W. et al. Understanding and modeling alternating tangential flow filtration for perfusion cell culture. Biotechnol. Prog. 30, 1291–1300 (2014).
Hong, J. et al. Development of an alternating tangential flow (ATF) perfusion-based transient gene expression (TGE) bioprocess for universal influenza vaccine. Biotechnol. Prog. 35, e2831 (2019).
Gollapudi, D. et al. Analytical characterization of broadly neutralizing antibody CAP256LS heavy chain clipping during manufacturing development. Biotechnol. Prog. https://doi.org/10.1002/btpr.3296 (2022).
Pan, Z. et al. Systematic analysis of the in situ crosstalk of tyrosine modifications reveals no additional natural selection on multiply modified residues. Sci. Rep. 4, 7331 (2014).
Julg, B. et al. Broadly neutralizing antibodies targeting the HIV-1 envelope V2 apex confer protection against a clade C SHIV challenge. Sci. Transl. Med. 9, 1321 (2017).
Zalevsky, J. et al. Enhanced antibody half-life improves in vivo activity. Nat. Biotechnol. 28, 157–159 (2010).
Gaudinski, M. R. et al. Safety and pharmacokinetics of the Fc-modified HIV-1 human monoclonal antibody VRC01LS: A phase 1 open-label clinical trial in healthy adults. PLoS Med. 15, e1002493 (2018).
Sarzotti-Kelsoe, M. et al. Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J. Immunol. Methods 409, 131–146 (2014).
Cai, C. X. et al. Tyrosine O-sulfation proteoforms affect HIV-1 monoclonal antibody potency. Sci. Rep. 12, 8433 (2022).
Gorman, J. et al. Structure of super-potent antibody CAP256-VRC26.25 in complex with HIV-1 envelope reveals a combined mode of trimer-apex recognition. Cell Rep. 31, 107488 (2020).
Avery, L. B. et al. Utility of a human FcRn transgenic mouse model in drug discovery for early assessment and prediction of human pharmacokinetics of monoclonal antibodies. MAbs 8, 1064–1078 (2016).
Rudicell, R. S. et al. Enhanced potency of a broadly neutralizing HIV-1 antibody in vitro improves protection against lentiviral infection in vivo. J. Virol. 88, 12669–12682 (2014).
Kwon, Y. D. et al. A matrix of structure-based designs yields improved VRC01-class antibodies for HIV-1 therapy and prevention. MAbs 13, 1946918 (2021).
Vlasak, J. & Ionescu, R. Fragmentation of monoclonal antibodies. MAbs 3, 253–263 (2011).
Gaudinski, M. R. et al. Safety and pharmacokinetics of broadly neutralising human monoclonal antibody VRC07-523LS in healthy adults: A phase 1 dose-escalation clinical trial. Lancet HIV 6, e667–e679 (2019).
Mahomed, S. et al. Assessing the safety and pharmacokinetics of the anti-HIV monoclonal antibody CAP256V2LS alone and in combination with VRC07-523LS and PGT121 in South African women: Study protocol for the first-in-human CAPRISA 012B phase I clinical trial. BMJ Open 10, e042247 (2020).
Kong, R. et al. Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody. Science 352, 828–833 (2016).
Bhiman, J. N. et al. Viral variants that initiate and drive maturation of V1V2-directed HIV-1 broadly neutralizing antibodies. Nat. Med. 21, 1332–1336 (2015).
Acknowledgements
The authors thank Cindy Cai for technical assistance, and Brenda Hartman for assistance with figures and manuscript preparation. They thank J. Baalwa, D. Ellenberger, F. Gao, B. Hahn, K. Hong, J. Kim, F. McCutchan, D. Montefiori, L. Morris, J. Overbaugh, E. Sanders-Buell, G. Shaw, R. Swanstrom, M. Thomson, S. Tovanabutra, C. Williamson and L. Zhang for contributing the HIV-1 Envelope plasmids used in our neutralization panel. The master cell bank for production of PGT121.414.LS was provided by Beth Israel Deaconess Medical Center. Funding was provided by the Intramural Research Programs of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, USA.
Funding
Open Access funding provided by the National Institutes of Health (NIH).
Author information
Authors and Affiliations
Contributions
B.Z., D.G., J.G., A.P., N.D.R. wrote the manuscript. J.G., A.P., M.A., K.M., C.H.S., N.D.R. prepared figures. B.Z., S.O., L.D., M.A., K.M., E.S.Y., A.P., B.L., L.G., W.H.L., S.D.S., J.S., M.S. performed experiments. C.C., X.C., C.L., W.S. provided unique reagents, L.G. and J.S. contributed critical information on reagents, V.I. developed methods, and M.L. curated data. A.M., K.C., J.M., P.D.K., Q.L., and N.D.R. supervised investigation and administration. All co-authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
NDR, JG, PDK, and JRM are co-inventors on a patent including the CAP256-VRC26.25 antibody, US application 16/718,322 pending. All other authors report no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, B., Gollapudi, D., Gorman, J. et al. Engineering of HIV-1 neutralizing antibody CAP256V2LS for manufacturability and improved half life. Sci Rep 12, 17876 (2022). https://doi.org/10.1038/s41598-022-22435-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-22435-2
- Springer Nature Limited