Abstract
Two new classes of hybrid quinoline–imidazole/benzimidazole derivatives (the hybrid QIBS salts and QIBC cycloadducts) were designed and synthesized to evaluate their anticancer and antimicrobial activity. The strategy adopted for synthesis is straight and efficient, in four steps: N-acylation, N-alkylation, quaternization and a Huisgen 3 + 2 cycloaddition. The in vitro single-dose anticancer assay of forty six hybrid quinoline-benzimidazole compounds reveal that one QIBS salt (11h), has an excellent quasi nonselective activity against all type of cancer cell with an excellent PGI in the area of 90–100% and very good lethality. Three others quinoline–imidazole/benzimidazole hybrids (8h, 12h, 12f) has an excellent selective activity against some cancer cell lines: breast cancer MDA-MB-468 and Leukemia HL-60 TB). The five-dose assay screening confirms that compound 11h possesses excellent anti-proliferative activity, with GI50 in the range of nano-molar, against some cancer cell lines: Leukemia HL-60 TB, Leukemia K-526, Leukemia RPMI-8226, Breast cancer MDA-MB-468, Lung cancer HOP-92 and Ovarian cancer IGROV1. The antibacterial assay indicates that three hybrid QIBS salts (12f, 12c, 12d) have an excellent activity against Gram-negative bacteria E. coli (superior to control Gentamicin) while against Gram-positive bacteria S. aureus only one compound 8i (R2 = -CF3) exhibits a significant activity (superior to control Gentamicin). The MIC assay indicates that two other compounds (11h, 12h) are biologically active to a very low concentration, in the range of nano-molar. We believe that all these excellent assets related to anticancer and antibacterial activities, make from our hybrid quinoline–imidazole/benzimidazole compounds bearing a phenyl group (R2 = –C6H5) in the para (4)-position of the benzoyl moiety a good candidate for future drug developing.
Similar content being viewed by others
Introduction
Cancer is one of the most merciless, serious and life-threatening disease worldwide, being characterized by the uncontrolled and rapid pathological growth of abnormal cells in the body and spread in different organs (metastasis)1. Based on WHO data, cancer continues to grow all around the world becoming the second leading cause of death worldwide (with over 10 million deaths in 2020) and exerting a great pressure on health systems, individuals and communities2,3,4,5.
Infectious diseases caused by microorganisms (especially bacteria and fungi) represent another major threat endanger human life and health6. In particular, overconsumption, widespread use and misuse of antimicrobials2,6 have caused seriously problems in the treatment of many microbial illnesses7,8.
In spite of the greatest advances achieved in both cancer and microbial therapy, the existing treatment suffers from some major limitations, these including drug resistance, multi-drug resistance, extensively-drug-resistance, often high toxicity levels and non specificity of drugs, high prices, etc.7,8,9,10,11,12. Thus, continued searching for newer and better anticancer and antimicrobial drugs remains a very important clue in medicinal chemistry.
A literature survey revealed that among heterocycles, quinoline and imidazole/benzimidazole derivatives are privileged scaffolds for the development of new drug entities. In this respect they possess a wide range of biological activities such as anticancer, antiplasmodial and antimalarial, antitubercular, antibacterial, antifungal, antiviral, anthelmintic, anti-HIV, analgesic, anticonvulsant, anti-inflammatory, antihistaminic, antipsychotic, anti-Alzheimer’s, antihypertensive, etc.9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26.
In continuation of our seeking for new entities with anticancer and antimicrobial activity with quinoline and imidazole/benzimidazole skeleton21,22,23,24,25,26, we report herein the design, synthesis and biological activity of two new class of hybrid molecules with quinoline–imidazole/benzimidazole skeleton.
Results and discussion
Design and synthesis
During the last few years molecular hybridization became a powerful tool in drug design and discovery offering an attractive approach to obtain better drugs for the treatment of a large variety of human diseases including cancer and microbial illness. One of the methods used for the construction of hybrid molecules combines two or more drug pharmacophores in a single multi-functional molecule using a linker chain. The main goals of this pharmacophore merging approach consist in the interaction of the resulting molecule with dual or multiple targets, amplifying the biological activity and specificity, reducing the known side effects associated with each hybrid part, reducing the drug-drug interactions26,27,28,29,30,31,32. By doing so, the hybrid drugs became more powerful and valuable than conventional classic drugs.
In previous research work we successfully identify quinoline and imidazole/benzimidazole derivatives with anticancer and antimicrobial activity22,23,24,25,26. Using molecular hybridization and our previous results in the field, in the present study we aimed to obtain two new classes of hybrid molecules which combine the quinoline and imidazole/benzimidazole pharmacophores, linked by an amide-alkyl unit. A first class of merged molecule has a quinoline–imidazole/benzimidazole salt structure (I, QIBS) while the second one has a quinoline–imidazole/benzimidazole cycloadduct structure (II, QIBC), Fig. 1. For both classes of molecules, the two pharmacophores will assure the anticancer and antimicrobial activity while the amide-alkyl linker/spacer will realize amide bonds and a different spatial orientation (via the different length of alkyl chain) between the quinoline–imidazole/benzimidazole units. The amide-alkyl linker/spacer will be also responsible for better pharmacological properties (especially water solubility and membrane permeability) and for a more suitable interaction with the putative binding site.
In the case of QIBS class I, the introduction of a nitrogen positive charge on imidazole moiety will increase membrane permeability and water solubility, and also will lead to more potent antibacterial compounds with broadened spectrum. In equal measure we were interested to study the influence of substituents R1 and R2 from the benzoyl moieties anchored to imidazole/benzimidazole unit. The literature data reveal that a halogen atom or a phenyl ring anchored on the benzoyl moiety, especially at the para position of the ring, have a positive influence on both antimicrobial and anticancer activities24,33,34,35.
Having in view the molecular hybridization approach used and our previously results24,35, we estimate as possible target (binding site) for our hybrid quinoline–imidazole/benzimidazole derivatives ATP synthase and Topoisomerase II. Also the literature data indicate zinc binding groups, amide and nitrogen from quinoline moiety, as a classic pharmacophoric model of histone deacetylases (HDAC) inhibitors36, Fig. 2.
As a matter of fact, the ability of amide group and nitrogen from quinoline moiety to bind zinc, is well known and experimentally proved by obtaining and fully characterized (X-ray on monocrystal including) of such of complexes, by us including23,37.
In order to obtain our QIBS and QIBC derivatives we used a similar procedure reported previously by our group21. The reaction pathway is straight and efficient, involving four steps indicated in Fig. 3. The initial N-acylation reaction of 8-aminoquinoline, is followed by an N-alkylation of the -NH- amino group from imidazole/ benzimidazole moiety leading to the key quinoline—imidazole/ benzimidazole intermediates, 4, 5, 9, 10. In the next step, a quaternization reaction of N-imidazole atom with variously activated halogenated derivatives 6a–k (ϖ–bromo/chloro 2,4-R1,R2-substituted-acetophenones), is leading to a first class of hybrid quinoline–imidazole/benzimidazole derivatives: the salts QIBS derived from imidazole 7a–k (with one methylene group as linker) and 8a–k (with two methylene groups as linker), respectively the salts QIBS derived from benzimidazole 11a–k (with one CH2 group) and 12a–k (with two CH2 groups). In the last step imidazolium and benzimidazolium ylides, generated in situ from the corresponding QIBS salts 7a–k, 8a–k respectively 11a–k, were treated with the activated alkyne dimethyl acetylenedicarboxylate (DMAD), when a Huisgen 3 + 2 cycloaddition take place, generating the second class of hybrid quinoline–imidazole/benzimidazole derivatives, the cycloadducts QIBC 13a–k, Fig. 3. It should be noted that the cycloaddition reaction with DMAD take place only in the case of benzimidazolium ylides while in the case of imidazolium ylides the reactions do not occur, leading to decomposition products.
As in related cases23, under conventional thermal heating (TH) the yields were low to moderate and the reactions conditions relatively harsh. As a result we perform the synthesis under ultrasounds (US) irradiation and the obtained results were much better: higher yields with about 10–20% and a significant reduction in reaction time, from hours to minutes.
The structures of the newly compounds were proven by elemental (C, H, N) and spectroscopic analysis: IR, 1H-NMR, 13C-NMR, and two-dimensional experiments 2D-COSY, 2D-HMQC, 2D-HMBC.
If we consider the most active and promising anticancer and antimicrobial compounds as representative for their series (n = 2, namely 8a, 8b, 8c, 8d, 8h and 12a, 12b, 12c, 12d, 12h), the most informative signals furnished by 1H-NMR spectra are those one of the protons from the three aliphatic methylene groups [H11, H12 (the ethylene linker), H1′ (from CH2–CO)], the aromatic protons H14 (from 2 position of imidazole ring), H4′, H5′ and protons from 6′ position (from benzoyl), and proton H9 from the amide functional group, Table 1.
The most deshielded protons are H9 (10.40–10.36 ppm, singlet) from amide functional group, which is characteristic for this functionality. The next deshielded protons are H14 (9.82–9.20 ppm, singlet) from the two positions of imidazole/benzimidazole moiety, due to the powerful deshielding effect of the positive nitrogen atom N15 and to N13 nitrogen. The protons from benzoyl moiety (H4′, H5′) appear at chemical shifts in accordance with the electronic effects exerted by the substituent from the para position of the benzoyl ring (8.46–7.14 ppm, doublet, J = 8.0 Hz). The signals of the H1′ protons from methylene group appear as a singlet, at a very low field, unusual for this type of protons, around 6.47–6.04 ppm. This is due to the powerful electron withdrawing effect of the adjacent carbonyl group and positive nitrogen atom N15 from imidazole/benzimidazole moiety. The signals of the protons from ethylene linker appear at 5.00–4.64 ppm for H12 (doublet, J = 6.5 Hz) respectively 3.47–3.34 ppm for H11 (triplet, J = 6.5 Hz), in accordance with the influence of their substituents: α-nitrogen atom N13, β-carbonyl group for H12, respective β-nitrogen atom N13, α-carbonyl group for H11. If we compare the spectra of compounds 12 (containing a benzimidazole moiety) with spectra of compounds 8 (containing a imidazole moiety), we may notice a clear influence of benzimidazole moiety related to the protons from ethylene linker (H11, H12) which consist in a deshielding process with about 0.35 ppm on H12 protons and 0.1 ppm on H11 protons, in accordance with the withdrawing effect of the heterocyclic ring.
In the 13C-NMR spectra of compounds 8 and 12, the most important data are furnished by the signals corresponding to the carbonyl groups (C10 and C2′), the three carbons from the methylene groups (C11, C12, C1′), the aromatic carbons C14 (from imidazole ring) and C3′, C4′, C5′, C6′ (from benzoyl moiety), Table 2.
The most deshielded signals are those one of the carbon from carbonyl ketone group (C2′) which appear around 190.7–189.4 ppm, typical for an alkyl-aryl C=O carbonyl ketone group. They are followed by the signals of the carbon from carbonyl amide group (C10, appear around 168 ppm), which is also typical for C=O carbonyl amide group. The heteroaromatic carbons C14 from imidazole ring appear around 144 ppm in compounds 12 (containing a benzimidazole moiety) respectively 137 ppm in compounds 8 (containing a imidazole moiety), in accordance with the powerful deshielding effect of the two adjacent nitrogen atoms from imidazole ring. The three carbons from the methylene groups (C11, C12, C1′) appear around 55–53 ppm (C1′: α-ketone, α-positive nitrogen), 45–43 ppm (C12: α-nitrogen atom N13, β-carbonyl group), 36–35 ppm (C11: β-nitrogen atom N13, α-carbonyl group).
The aromatic carbons from benzoyl moiety (C3′, C4′, C5′, C6′) appear at chemical shifts in accordance with the electronic effects exerted by the substituent from the para position of the benzoyl ring.
All the remaining signals from NMR spectra are in accordance with the proposed structures. See also Experimental part for the 1H- and 13C-NMR spectra for compounds 8a, 8b, 8c, 8d, 8h, 12a, 12b, 12c, 12d, 12h.
Anticancer activity
The in vitro anticancer assay was performed at the National Cancer Institute (NCI, USA), under the Developmental Therapeutics Program (DTP). The 60 created cell lines represent nine human cancers: breast, central nervous system, colon, kidney, leukemia, lung, melanoma, ovary, and prostate. The cancer cell lines grewup according to the NCI standard protocols. The DTP screens include the NCI 60 cell line screen, utilizing 60 different human tumor cell lines in accordance with the protocol of the NCI38,39,40. The screening is beginning with the evaluation of all compounds against the 60 cell lines at a single-dose of 10 μM38. The output from the single dose screen is reported as a mean graph and is available for analysis by the COMPARE program (See: http://dtp.nci.nih.gov/docs/compare/compare.html). The results are expressed as "Percentage Growth Inhibition" (PGI) term, and represent growth relative to the no-drug control, and relative to the time zero number of cells. This allows detection of both growth inhibition (values between 0 and 100) and lethality (values less than 0). For example, a value of 40 would mean 60% growth inhibition while a value of -40 would mean 40% lethality (more information could be found to Supporting Information data of this article).
Forty six of the synthesized compounds were selected and tested by the NCI for the primary single dose assay (10–5 M), the obtained results being presented at Supporting Information data (in Supplementary Table 1 (QIBS salts, 7a, b, c, d, e, f, h, k and 11a, b, c, d, e, f, g, h, k), Supplementary Table 2 (QIBS salts, 8a, b, c, d, e, f, g, h, i, j, k), Supplementary Table 3 (QIBS salts, 12a, b, c, d, e, f, g, h, i, j, k), Supplementary Table 4 (QIBC cycloadducts 13a, c, d, e, f, g, h). In Table 3 are summarized the results of the NCI 60 anticancer primary single-dose assay for the most active compounds.
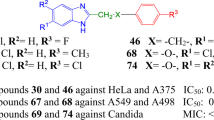
From Table 3 we may notice the most active compound is QIBS salt, 11h (hybrid quinoline-benzimidazole salt, n = 1), with a quasi nonselective activity against all type of cancer cell (except renal and prostate) with an excellent PGI in the area of 90–100% and very good lethality. Another excellent result is the selectivity against some cancer cell lines exerted by some hybrid quinoline-imidazole/benzimidazole salts with ethylene linker (n = 2): 12h on two cancer lines [Breast cancer MDA-MB-468 (PGI = 100% and L = 32%) and Leukemia HL-60 TB (PGI = 100% and L = 42%)], respectively to one cancer line 12f [Breast cancer MDA-MB-468 (PGI = 100% and L = 17%)] and 8h [Breast cancer MDA-MB-468 (PGI = 100% and L = 26%)].
The results obtained for the hybrid quinoline–imidazole/benzimidazole cycloadducts (13a, c, d, e, f, g, h) were disappointing, with low value of PGI (see Table 4 from Supporting Information data).
Compound 11h which had the best growth inhibition profile among the tested compounds, was selected by NCI for detailed studies at five different concentrations (0.01, 0.1, 1, 10 and 100 μM) in the five-dose assay studies38,39,40, selected results being presented in Table 4.
The in vitro five-dose assay screening results revealed that compound 11h possesses excellent anti-proliferative activity, with GI50 in the range of nano-molar, against some cancer cell lines: Leukemia HL-60 TB (428 nM), Leukemia K-526 (639 nM), Leukemia RPMI-8226 (911 nM), Breast cancer MDA-MB-468 (590 nM), Lung cancer HOP-92 (677 nM) and Ovarian cancer IGROV1 (785 nM). Compound 11h possesses also a good to moderate activity with GI50 values ranging from 1 to 3 μM against the remaining cancer cells from all nine sub-panels.
From SAR point of view, the following remarks could be done for hybrid quinoline–imidazole/benzimidazole compounds with anticancer activity:
-
the most important factor that influence the biological activity is the anchored substituent from the para (4)-position of the benzoyl moiety, compounds with a phenyl (–C6H5) group having the highest and the most interesting activity. The presence of a fluoro (–F), nitro (–NO2), and methoxy (–OCH3) moiety are also useful for activity;
-
the hybrid compounds with benzimidazole moiety (11 and 12) are more active than those one with a imidazole structure (7 and 8);
-
the length of alkyl chain linker betwen quinoline and imidazole/benzimidazole moieties, influence selectivity of compounds: increasing size of alkyl length (from one methylene to two methylene groups) increase substantially the selectivity. Thus, while the benzimidazole compound 11h (with one methylene, n = 1) have a high quasi nonselective anticancer activity, the benzimidazole compound 12h (with two methylene, n = 2) have a high selective anticancer activity. We presume that this behaviour is related to the interaction with the receptor situs;
-
the QIBS salts compounds are more active than QIBC cycloadducts, which confirms our initial hypothesis that the introducing of a nitrogen positive charge on imidazole moiety will increase membrane permeability and water solubility, and also will lead to more potent biologically active compounds.
Antimicrobial activity
The in vitro antibacterial and antifungal activities of the hybrid quinoline–imidazole/benzimidazole compounds was determined by the Kirby-Bauer disk diffusion method41. The method is using μler Hinton nutrient agar medium for antibacterial assay and Sabouraud nutrient agar medium for antifungal assay. The in vitro antibacterial activity was evaluated against Gram-positive bacteria Staphylococcus aureus ATCC 25923 and Gram-negative Escherichia coli ATCC 25922 and the antifungal activity against fungus Candida albicans ATCC 10231. Gentamicin was used as positive control (C+) for S. aureus and E. coli and Nystatin for C. albicans. The negative control (C−) consists of sterile filter paper disks (with no antimicrobial compounds) inoculated with DMSO 3%. The obtained results are expressed as diameters of inhibition zones (mm), the larger diameter is, the most active compounds are. The obtained results are presented at Supporting Information data (in Supplementary Tables 5–8). In Table 5 below are summarized the results of the antibacterial and antifungal assay for the most active compounds.
The data from Table 5 reveals that some of our hybrid quinoline–imidazole/benzimidazole compounds proved to be effective against the tested bacterial strains. Our hybrid compounds manifest a much pronounced antibacterial activity against gram-negative bacteria E. coli comparative with gram-positive bacteria S. aureus, three compounds (12f, 12c, 12d) being extremely active (having a diameter of zone inhibition up to 20 mm, superior to control Gentamicin with about 8–12 mm), while seven others (11f, 11d, 8h, 8d, 7j, 7i, 7d) are very active (having a diameter of zone inhibition superior to control Gentamicin with more than 5 mm). Against gram-positive bacteria S. aureus only one compound 8i (R2 = − CF3) manifest a significant activity, superior to control Gentamicin with 6 mm.
Against fungus C. albicans the tested hybrid quinoline–imidazole/benzimidazole derivatives proved to be inactive.
From SAR point of view, the following remarks could be done for hybrid quinoline–imidazole/benzimidazole compounds with antibacterial activity:
-
presence of a hybrid quinoline–imidazole/benzimidazole moiety have a beneficial influence for antibacterial activity against gram-negative bacteria E. coli, lesser influence against gram-positive bacteria S. aureus and no influence against fungus;
-
the most important factor that influence the biological activity within the hybrid quinoline–imidazole/benzimidazole derivatives, is the presence of a benzimidazole moiety into the hybrid structure;
-
another important factor that influence the antibacterial activity is the anchored substituent from the para (4)-position of the benzoyl moiety. The presence of classical isosteres atoms/groups fluoro (–F), chloro (–Cl) and methyl (–CH3) or nonclassical isosteres groups trifloromethyl (–CF3), are leading to very active compounds [12f (R2 = –F), 12c (R2 = –Cl), 12d (R2 = –Me)] respectively to active compounds [11f (R2 = –F), 11d (R2 = –Me), 8i (R2 = –CF3), 8d (R2 = –Me), 7i (R2 = –CF3), 7d (R2 = –Me)].
-
the length of alkyl chain linker between quinoline and imidazole/benzimidazole moieties, seems to have no significant influence on activity.
In the next step of antimicrobial assay, the minimum inhibitory concentration (MIC) of the hybrid quinoline–imidazole/benzimidazole compounds were determined, using the standardized broth microdilution assay procedure42,43,44,45,46. The resulted MIC value is defined as the lowest concentration of the antimicrobial compounds under investigation, which prevents visible growth of the tested microorganism. The obtained results are presented at Supporting Information data (in Tables 9–12). In Table 6 below are summarized the results of the antibacterial and antifungal MIC assay for the most active compounds.
The data from Table 6, indicate that two hybrid compounds (with phenyl group (R2 = –C6H5) in the para (4)-position of the benzoyl moiety) are active to a very low concentration, in the range of nano-molar against Staphylococcus aureus, having a MIC of 0.01 ng/mL in the case of 11h, respectively 0.3 ng/mL in the case of 12h. These two hybrid compounds are also active to a low concentration against Escherichia coli, in the range of micro-molar, having a MIC of 0.004 µg/mL each. Significant results were obtained against S. aureus for compounds 11g and 11i (with a MIC of 0.009 µg/mL) and against E. coli for compounds 11f and 7h (with a MIC of 0.009 µg/mL respectively 0.001 µg/mL).
Conclusions
In summary, we report herein the design, synthesis, anticancer and antimicrobial activity of two new classes of hybrid quinoline–imidazole/benzimidazole derivatives: the salts QIBS (derived from imidazole and benzimidazole) respectively the cycloadducts QIBC. The reaction pathway is straight and efficient, involving four steps: N-acylation, N-alkylation, quaternization and a Huisgen 3 + 2 cycloaddition. Forty six of the synthesized compounds were selected and tested by the NCI for the in vitro primary single dose anticancer assay and one compound for the five-dose assay. One hybrid quinoline-benzimidazole salt, 11h, has an excellent quasi nonselective activity against all type of cancer cell (except renal and prostate) with an excellent PGI in the area of 90–100% and very good lethality. Three other quinoline–imidazole/benzimidazole hybrids has an excellent selective activity against some cancer cell lines: 12h on two cancer lines (Breast cancer MDA-MB-468 and Leukemia HL-60TB), 12f on Breast cancer MDA-MB-468 and 8h also on Breast cancer MDA-MB-468. The five-dose assay screening confirm that compound 11h possesses excellent anti-proliferative activity, with GI50 in the range of nano-molar, against some cancer cell lines: Leukemia HL-60 TB, Leukemia K-526, Leukemia RPMI-8226, Breast cancer MDA-MB-468, Lung cancer HOP-92 and Ovarian cancer IGROV1. The SAR correlation related to anticancer activity reveals interesting data related to the influence of heterocycle (a benzimidazole moiety is favourable), length of alkyl chain linker (increasing the size of alkyl length affect substantially the selectivity) and substituent from the para (4)-position of the benzoyl moiety, compounds with a phenyl (–C6H5) group having the highest and the most interesting activity. Overall, the hybrid QIBS salts are more active than QIBC cycloadducts. Three hybrid QIBS salts (12f, 12c, 12d) have an excellent antibacterial activity against gram-negative bacteria E. coli (superior to control Gentamicin). Against gram-positive bacteria S. aureus one compound 8i (R2 = –CF3) manifest a significant activity (superior to control Gentamicin) and the MIC assay indicate that two other compounds (11h, 12h) are active to a very low concentration, in the range of nano-molar. Overall, the hybrid QIBS salts manifest a much pronounced antibacterial activity against gram-negative bacteria E. coli comparative with gram-positive bacteria S. aureus. Against fungus C. albicans the tested hybrid quinoline–imidazole/benzimidazole derivatives are inactive. The SAR correlation related to antibacterial activity reveals interesting data, the presence of a benzimidazole moiety being favourable for activity, the length of alkyl chain linker has no influence, while the presence of isosteres fluoro (–F), chloro (–Cl), methyl (–CH3) or trifloromethyl (–CF3) anchored in para (4-) position of the benzoyl moiety increase substantially the activity. We believe that all these excellent assets related to anticancer and antibacterial activity, make from our hybrid quinoline–imidazole/benzimidazole compounds bearing a phenyl group (R2 = –C6H5) in the para (4)-position of the benzoyl moiety a good candidate for future drug developing.
Experimental
Materials and measurements
Melting points were determined using an electrothermal Mel-Temp apparatus and were uncorrected. The NMR spectra were recorded on a Bruker Avance III 500 MHz spectrometer operating at 500 MHz for 1H and 125 MHz for 13C. The NMR apparatus is equipped with a 5 mm PABBO detection probe, and the program used for acquisition and processing data is TopSpin 3.2 PL5. The following abbreviations were used to designate chemical shift multiplicities: s = singlet, bs = broad singlet, d = doublet, dd = doublet of doublets, ad = apparent doublet, add = apparent doublet of doublets, t = triplet, at = apparent triplet, m = multiplet. Chemical shifts were reported in δ units (ppm) relative to the residual peaks of solvents (ref: DMSO, 1H: 2.50 ppm; 13C: 39.52 ppm or CDCl3, 1H: 7.26 ppm; 13C: 77.16 ppm. Coupling constants (J) were given in Hz. Infrared (IR) data were recorded as films on potassium bromide (KBr) pellets on a FT-IR VERTEX 70 Bruker spectrophotometer. The microanalyses were in satisfactory agreement with the calculated values: C, ± 0.15; H, ± 0.10; N, ± 0.30. Ultrasound assisted reactions were accomplished using Sonics (Sonics VCX-130, USA), with a nominal power of 130 W and a frequency of 20 kHz. For this ultrasonic reactor the titanium horn (diameter: 6 mm; length: 116 mm) was fixed firmly to the ultrasonic converter. The titanium probe tip was directly immersed in the used solvent. Thin layer chromatography (TLC) was carried out on Merck silica gel 60 F254 plates. Visualisation of the plates was achieved using a UV lamp (λmax = 254 or 365 nm). The ultrasonic bath Elma Transsonic T310 (power 34.5 W, frequency 35 kHz) was used for solubilising the starting materials.
Compounds 2, 3, 4, 7a–k, 9, 11a–k and 13a–k were previous reported21.
General procedure for the synthesis of starting materials 5 and 10
Imidazole (5.5 mmol)/benzimidazole (5.5 mmol) are solubilized in 20 mL anhydrous acetonitrile (ACN) and NaH 60% is gradually added (15 mmol, 0.6 g), suspended in anhydrous ACN, this being pre-washed with n-hexane to remove the mineral oil. To the resulting mixture was added the corresponding propionamide 3 (5 mmol), solubilized in 25 mL of anhydrous ACN. After addition of the reagents, the reaction is continued for 8 h at room temperature (TLC monitoring). Reaction processing consisted of simple filtration of the precipitate formed, and the solution in the flask was concentrated on a rotaevaporator. The precipitate obtained is purified by separation on a chromatographic column, using silica gel as a solid stationary phase, and a mixture of CH2Cl2 and CH3OH (96/4) as eluent.
General procedure for the synthesis of quaternary salts 8a–k and 12a–k
The solution that contains parental hybrid compound 5 or 10 (1 mmol) and corresponding phenacyl bromide/chloride 6a–k (1.2 mmol) was placed in a reaction vessel and was exposed under US irradiation. The best results were obtained by applying a pulse irradiation (5 s pulse/5 s pause, 100% from the full power of the generator). Once the irradiation cycle was completed, the reaction vessel was removed and the obtained precipitate was separated by filtration and was washed with 5–7 mL of acetone. No other purification was required. The reactions take place under ultrasound irradiation between 20 and 100 min.
Spectral data of starting materials 5 and 10
Figure 4 shows the numbering of the starting compounds.
3-(1H-imidazol-1-yl)-N-(quinolin-8-yl)propanamide (5)
White powder, 42% yield, m.p. = 167–170 °C. 1H-NMR (500 MHz, DMSO-d6): δ = 3.14 (t, J = 6.5 Hz, 2H, H11), 4.32 (t, J = 6.5 Hz, 2H, H12), 6.85 (s, 1H, H16), 7.21 (s, 1H, H17), 7.57 (t, J = 8.0 Hz, 1H, H6), 7.62 (dd, J = 4.0 Hz, J = 8.0 Hz, 1H, H3), 7.67–7.66 (m, 2H, H5, H14), 8.39 (d, J = 8.0 Hz, 1H, H4), 8.62 (d, J = 7.5 Hz, 1H, H7), 8.92 (ad, J = 4.0 Hz, 1H, H2), 10.23 (s, 1H, H9). 13C-NMR (125 MHz, DMSO-d6): δ = 37.8, 42.2, 117.0, 119.3, 122.1, 126.9, 127.8, 128.3, 134.3, 136.5, 137.3, 138.1, 148.8, 169.2. Anal. Calcd. for C15H14N4O: C, 67.65; H, 5.30; N, 21.04; found: C, 67.75; H, 5.20; N, 21.24.
3-(1H-benzo[sd]imidazol-1-yl)-N-(quinolin-8-yl)propanamide (10)
White powder, 54% yield, m.p. = 160–163 °C. 1H-NMR (500 MHz, CDCl3): δ = 3.12 (t, J = 6.5 Hz, 2H, H11), 4.70 (t, J = 6.5 Hz, 2H, H12), 7.34–7.27 (m, 2H, H6, H18), 7.42 (dd, J = 4.5 Hz, J = 8.5 Hz, 1H, H3), 7.55–7.49 (m, 3H, H5, H17, H19), 7.79 (d, J = 8.0 Hz, 1H, H7), 8.07 (s, 1H, H14), 8.13 (add, J = 8.5 Hz, 1H, H4), 8.73–8.70 (m, 2H, H2, H16), 9.73 (bs, 1H, H9). 13C-NMR (125 MHz, CDCl3): δ = 37.6, 40.5, 109.5, 116.8, 120.6, 121.8, 122.1, 122.3, 123.1, 127.4, 128.0, 133.6, 133.9, 136.5, 138.2, 143.7, 144.0, 148.3, 167.9. Anal. Calcd. for C19H16N4O: C, 72.13; H, 5.10; N, 17.71; found: C, 72.03; H, 5.15; N, 17.61.
Spectral data of quaternary salts 8a–k and 12a–k
Figure 5 shows the numbering of the quaternary salts.
3-(2-(4-Nitrophenyl)-2-oxoethyl)-1-(3-oxo-3-(quinolin-8-ylamino)propyl)-1H-imidazol-3-ium bromide (8a)
Yellow powder, 80% yield; m.p. = 190–192 °C. 1H-NMR (500 MHz, DMSO-d6): δ = 3.34 (at, J = 6.5 Hz, 2H, H11), 4.65 (t, J = 6.5 Hz, 2H, H12), 6.13 (s, 2H, H1’), 7.58 (t, J = 8.0 Hz, 1H, H6), 7.65 (dd, J = 4.0 Hz, J = 8.0 Hz, 1H, H3), 7.72–7.69 (m, 2H, H5, H16), 7.94 (s, 1H, H17), 8.26 (d, J = 9.0 Hz, 2H, 2xH4’), 8.44–8.41 (m, 3H, H4, 2xH5’), 8.60 (d, J = 8.0 Hz, 1H, H7), 8.95 (add, J = 4.0 Hz, 1H, H2), 9.20 (s, 1H, H14), 10.37 (bs, 1H, H9). 13C-NMR (125 MHz, DMSO-d6): δ = 36.2, 45.3, 55.8, 117.3, 122.2, 122.3, 122.4, 123.9, 124.1, 126.8, 127.9, 129.6, 134.2, 136.6, 137.7, 138.2, 138.3, 148.9, 150.5, 168.6, 190.7. IR (KBr) v/cm-1: 3327, 3014, 2976, 1715, 1684, 1524, 1343, 1182. Anal. Calcd. for C23H20BrN5O4: C, 54.13; H, 3.95; N, 13.72; found: C, 54.18; H, 3.90; N, 13.62.
3-(2-(4-Methoxyphenyl)-2-oxoethyl)-1-(3-oxo-3-(quinolin-8-ylamino)propyl)-1H-imidazol-3-ium bromide (8b)
White powder, 87% yield; m.p. = 204–205 °C. 1H-NMR (500 MHz, DMSO-d6): δ = 3.34 (at, J = 7.0 Hz, 2H, H11), 3.89 (s, 3H, p-OCH3, 4.64 (t, J = 7.0 Hz, 2H, H12), 6.10 (s, 2H, H1’), 7.14 (d, J = 9.0 Hz, 2H, 2xH5’), 7.58 (t, J = 8.0 Hz, 1H, H6), 7.65 (dd, J = 4.0 Hz, J = 8.0 Hz, 1H, H3), 7.71–7.69 (m, 2H, H5, H17), 7.91 (s, 1H, H16), 8.01 (d, J = 9.0 Hz, 2H: 2xH4’), 8.42 (add, J = 8.0 Hz, 1H, H4), 8.59 (d, J = 8.0 Hz, 1H, H7), 8.95 (add, J = 4.0 Hz, 1H, H2), 9.20 (s, 1H, H14), 10.37 (s, 1H, H9). 13C-NMR (125 MHz, DMSO-d6): δ = 36.2, 45.2, 55.0, 55.7 (p-OCH3), 114.3, 117.3, 122.1, 122.2, 122.3, 124.0, 126.5, 126.8, 127.9, 130.5, 134.2, 136.6, 137.7, 138.2, 148.9, 164.0, 168.6, 189.5. IR (KBr) v/cm−1: 3325, 3018, 2960, 1678, 1603, 1560, 1230, 1162. Anal. Calcd. for C24H23BrN4O3: C, 58.19; H, 4.68; N, 11.31; found: C, 58.29; H, 4.78; N, 11.11.
3-(2-(4-Chlorophenyl)-2-oxoethyl)-1-(3-oxo-3-(quinolin-8-ylamino)propyl)-1H-imidazol-3-ium bromide (8c)
Beige powder, 83% yield; m.p. = 218–220 °C. 1H-NMR (500 MHz, DMSO-d6): δ = 3.35 (at, J = 6.5 Hz, 2H, H11), 4.65 (t, J = 6.5 Hz, 2H, H12), 6.09 (s, 2H, H1’), 7.58 (t, J = 8.0 Hz, 1H, H6), 7.64 (dd, J = 4.0 Hz, J = 8.0 Hz, 1H, H3), 7.73–7.69 (m, 4H, H5, 2xH5’, H16), 7.94 (s, 1H, H17), 8.05 (d, J = 9.0 Hz, 2H, 2xH4’), 8.42 (add, J = 8.0 Hz, 1H, H4), 8.59 (d, J = 8.0 Hz, 1H, H7), 8.95 (add, J = 4.0 Hz, 1H, H2), 9.22 (s, 1H, H14), 10.36 (s, 1H, H9). 13C-NMR (125 MHz, DMSO-d6): δ = 36.2, 45.2, 55.4, 117.3, 122.1, 122.3, 122.4, 124.0, 126.8, 127.9, 129.2, 130.0, 132.4, 134.2, 136.6, 137.7, 138.2, 139.3, 148.9, 168.6, 190.5. IR (KBr) v/cm-1: 3332, 3008, 2987, 1670, 1530, 732. Anal. Calcd. for C23H20BrClN4O2: C, 55.27; H, 4.03; N, 11.21; found: C, 55.37; H, 4.08; N, 11.06.
3-(2-Oxo-2-(p-tolyl)ethyl)-1-(3-oxo-3-(quinolin-8-ylamino)propyl)-1H-imidazol-3-ium bromide (8d)
White powder, 90% yield; m.p. = 195–198 °C. 1H-NMR (500 MHz, DMSO-d6): δ = 2.41 (s, 3H, p-CH3), 3.34 (at, J = 6.0 Hz, 2H, H11), 4.64 (t, J = 6.0 Hz, 2H, H12), 6.04 (s, 2H, H1’), 7.43 (d, J = 8.5 Hz, 2H, 2xH5’), 7.58 (t, J = 8.0 Hz, 1H, H6), 7.65 (dd, J = 4.0 Hz, J = 8.0 Hz, 1H, H3), 7.70 (d, J = 8.0 Hz, 1H, H5), 7.73 (bs, 1H, H16), 7.93 (ad, J = 8.5 Hz, 3H, 2xH4’, H17), 8.42 (add, J = 8.0 Hz, 1H, H4), 8.60 (d, J = 8.0 Hz, 1H, H7), 8.95 (add, J = 4.0 Hz, 1H, H2), 9.21 (s, 1H, H14), 10.37 (s, 1H, H9). 13C-NMR (125 MHz, DMSO-d6): δ = 21.3 (p-CH3), 36.2 (C11), 45.2 (C12), 55.2 (C1’), 117.3 (C7), 122.1 (C3), 122.2 (C5), 122.3 (C17), 124.0 (C16), 126.8 (C6), 127.9 (C4a), 128.2 (2xC4’), 129.6 (2xC5’), 131.2 (C3’), 134.2 (C8), 136.6 (C4), 137.7 (C14), 138.2 (C8a), 145.1 (C6’), 148.9 (C2), 168.6 (C10), 190.7 (C2’). IR (KBr) v/cm-1: 3262, 3035, 2993, 1745, 1686, 1541, 1238, 1184, 1074. Anal. Calcd. for C24H23BrN4O2: C, 60.13; H, 4.84; N, 11.69; found: C, 60.03; H, 4.74; N, 11.79.
3-(2-(4-Cyanophenyl)-2-oxoethyl)-1-(3-oxo-3-(quinolin-8-ylamino)propyl)-1H-imidazol-3-ium bromide (8e)
White powder, 80% yield; m.p. = 213–216 °C. 1H-NMR (500 MHz, DMSO-d6): δ = 3.34 (2H: H11, J = 6.0 Hz, at), 4.65 (2H: H12, J = 6.0 Hz, t), 6.11 (2H: H1’, s), 7.59 (1H: H6, J = 8.0 Hz, t), 7.65 (1H: H3, J = 4.0 Hz, J = 8.0 Hz, dd), 7.67 (1H: H5, J = 8.0 Hz, d), 7.72 (1H: H16, bs), 7.94 (1H: H17, bs), 8.12 (2H: 2xH5’, J = 8.0 Hz, d), 8.18 (2H: 2xH4’, J = 8.0 Hz, d), 8.42 (1H: H4, J = 8.0 Hz, d), 8.59 (1H: H7, J = 8.0 Hz, d), 8.95 (1H: H2, J = 4.0 Hz, ad), 9.20 (1H: H14, s), 10.37 (1H: H9, s). 13C-NMR (125 MHz, DMSO-d6): δ = 36.2, 45.2, 55.7, 116.1, 117.3, 117.9, 122.1, 122.3, 122.3, 123.9, 126.8, 127.9, 128.7, 133.0, 134.2, 136.6, 136.9, 137.7, 138.2, 148.9, 168.6, 190.9. IR (KBr) v/cm-1: 3325, 3026, 2973, 2234, 1725, 1530, 1230, 1078. Anal. Calcd. for C24H20BrN5O2: C, 58.79; H, 4.11; N, 14.28; found: C, 58.89; H, 4.01; N, 14.08.
3-(2-(4-Fluorophenyl)-2-oxoethyl)-1-(3-oxo-3-(quinolin-8-ylamino)propyl)-1H-imidazol-3-ium bromide (8f)
White powder, 83% yield; m.p. = 206–209 °C. 1H-NMR (500 MHz, DMSO-d6): δ = 3.35 (at, J = 6.5 Hz, 2H, H11), 4.65 (t, J = 6.5 Hz, 2H, H12), 6.08 (s, 2H, H1’), 7.47 (t, J = 8.5 Hz, 2H, 2xH5’), 7.58 (t, J = 8.0 Hz, 1H, H6), 7.65 (dd, J = 4.0 Hz, J = 8.0 Hz, 1H, H3), 7.70 (d, J = 8.0 Hz, 1H, H5), 7.73 (bs, 1H, H16), 7.93 (bs, 1H, H17), 8.13 (aq, J = 8.5 Hz, 2H, 2xH4’), 8.42 (add, J = 8.0 Hz, 1H, H4), 8.59 (d, J = 8.0 Hz, 1H, H7), 8.95 (ad, J = 4.0 Hz, 1H, H2), 9.22 (s, 1H, H14), 10.37 (s, 1H, H9). 13C-NMR (125 MHz, DMSO-d6): δ = 36.2, 45.2, 55.3, 116.2, 117.3, 122.1, 122.3, 122.4, 124.0, 126.8, 127.9, 130.5, 131.2, 134.2, 136.6, 137.7, 138.2, 148.9, 165.6, 168.6, 190.0. IR (KBr) v/cm−1: 3295, 3024, 2975, 1701, 1692, 1543, 1233, 1174. Anal. Calcd. for C23H20BrFN4O2: C, 57.15; H, 4.17; N, 11.59; found: C, 57.05; H, 4.12; N, 11.74.
3-(2-(4-Bromophenyl)-2-oxoethyl)-1-(3-oxo-3-(quinolin-8-ylamino)propyl)-1H-imidazol-3-ium bromide (8g)
White powder, 85% yield; m.p. = 189–192 °C. 1H-NMR (500 MHz, DMSO-d6): δ = 3.35 (at, J = 6.5 Hz, 2H, H11), 4.64 (t, J = 6.5 Hz, 2H, H12), 6.04 (s, 2H, H1’), 7.58 (t, J = 8.0 Hz,1H, H6), 7.65 (dd, J = 4.5 Hz, J = 8.5 Hz, 1H, H3), 7.70–7.69 (m, 2H, H5, H16), 7.86 (d, J = 8.5 Hz, 2H, 2xH5’), 7.92 (bs, 1H, H17), 7.96 (d, J = 8.5 Hz, 2H, 2xH4’), 8.42 (d, J = 8.5 Hz, 1H, H4), 8.59 (d, J = 8.0 Hz, 1H, H7), 8.95 (ad, J = 4.5 Hz, 1H, H2), 9.18 (s, 1H, H14), 10.37 (s, 1H, H9). 13C-NMR (125 MHz, DMSO-d6): δ = 36.2, 45.2, 55.3, 117.3, 122.2, 122.3, 122.4, 124.0, 126.8, 127.9, 128.6, 130.1, 132.2, 132.7, 134.2, 136.6, 137.7, 138.2, 148.9, 168.6, 190.7. IR (KBr) v/cm−1: 3241, 3027, 2986, 1742, 1702, 1529, 1223, 1175, 681. Anal. Calcd. for C23H20Br2N4O2: C, 50.76; H, 3.70; N, 10.29; found: C, 50.66; H, 3.65; N, 10.44.
3-(2-([1,1′-Biphenyl]-4-yl)-2-oxoethyl)-1-(3-oxo-3-(quinolin-8-ylamino)propyl)-1H-imidazol-3-ium bromide (8h)
White powder, 86% yield; m.p. = 211–214 °C. 1H-NMR (500 MHz, DMSO-d6): δ = 3.36 (at, J = 6.5 Hz, 2H, H11), 4.66 (t, J = 6.5 Hz, 2H, H12), 6.11 (s, 2H, H1’), 7.46 (t, J = 7.5 Hz, 1H, H10’), 7.53 (t, J = 7.5 Hz, 2H, 2xH9’), 7.59 (t, J = 8.0 Hz, 1H, H6), 7.65 (dd, J = 4.0 Hz, J = 8.0 Hz, 1H, H3) 7.70 (d, J = 8.0 Hz, 1H, H5), 7.74 (bs, 1H, H16), 7.80 (d, J = 7.5 Hz, 2H, 2xH8’), 7.94 (ad, J = 8.5 Hz, 3H, 2xH5’, H17), 8.12 (d, J = 8.5 Hz, 2H, 2xH4’), 8.43 (d, J = 8.0 Hz, 1H, H4), 8.61 (d, J = 8.0 Hz, 1H, H7), 8.95 (ad, J = 4.0 Hz, 1H, H2), 9.23 (s, 1H, H14), 10.38 (s, 1H, H9). 13C-NMR (125 MHz, DMSO-d6): δ = 36.2, 45.2, 55.4, 117.3, 122.2, 122.3, 122.4, 124.0, 126.8, 127.1, 127.1, 127.9, 128.7, 128.9, 129.1, 132.5, 134.2, 136.6, 137.8, 138.2, 138.5, 145.7, 148.9, 168.6, 190.8. IR (KBr) v/cm−1: 3238, 3015, 2986, 1698, 1680, 1575, 1215. Anal. Calcd. for C29H25BrN4O2: C, 64.33; H, 4.65; N, 10.35; found: C, 64.43; H, 4.60; N, 10.25.
3-(2-Oxo-2-(4-(trifluoromethyl)phenyl)ethyl)-1-(3-oxo-3-(quinolin-8-ylamino)propyl)-1H-imidazol-3-ium bromide (8i)
White powder, 79% yield; m.p. = 219–221 °C. 1H-NMR (500 MHz, DMSO-d6): δ = 3.35 (at, J = 6.5 Hz, 2H, H11), 4.65 (t, J = 6.5 Hz, 2H, H12), 6.11 (s, 2H, H1’), 7.59 (t, J = 8.0 Hz, 1H, H6), 7.65 (dd, J = 4.5 Hz, J = 8.5 Hz, 1H, H3), 7.71–7.69 (m, 2H, H5, H16), 7.93 (bs, 1H, H17), 8.02 (d, J = 8.5 Hz, 2H, 2xH5’), 8.23 (d, J = 8.5 Hz, 2H, 2xH4’), 8.42 (add, J = 8.5 Hz, 1H, H4,), 8.60 (d, J = 8.0 Hz, 1H, H7), 8.95 (add, J = 4.5 Hz, 1H, H2), 9.19 (s, 1H, H14), 10.38 (s, 1H, H9,). 13C-NMR (125 MHz, DMSO-d6): δ = 36.2, 45.3, 55.6, 117.3, 122.2, 122.3, 122.4, 123.6, 124.0, 126.1, 126.9, 127.9, 129.0, 133.5, 134.2, 136.6, 136.9, 137.7, 138.2, 148.9, 168.6, 191.0. IR (KBr) v/cm−1: 3312, 3021, 2983, 1680, 1586, 1238, 1165. Anal. Calcd. for C24H20BrF3N4O2: C, 54.05; H, 3.78; N, 10.50; found: C, 54.15; H, 3.73; N, 10.40.
3-(2-Oxo-2-phenylethyl)-1-(3-oxo-3-(quinolin-8-ylamino)propyl)-1H-imidazol-3-ium bromide (8j)
White powder, 82% yield; m.p. = 197–201 °C. 1H-NMR (500 MHz, DMSO-d6): δ = 3.35 (at, J = 6.5 Hz, 2H, H11), 4.65 (t, J = 6.5 Hz, 2H, H12), 6.08 (s, 2H, H1’), 7.76–7.57 (m, 7H, H3, H5, H6, 2xH5’, H6’, H16), 7.93 (bs, 1H, H17), 8.04 (d, J = 7.0 Hz, 2H, 2xH4’), 8.42 (add, J = 8.5 Hz, 1H, H4), 8.60 (d, J = 8.0 Hz, 1H, H7), 8.95 (add, J = 4.0 Hz, 1H, H2), 9.21 (s, 1H, H14), 10.38 (s, 1H, H9). 13C-NMR (125 MHz, DMSO-d6): δ = 36.2, 45.2, 55.4, 117.3, 122.2, 122.3, 122.4, 124.0, 126.8, 127.9, 128.1, 129.1, 133.6, 134.2, 134.5, 136.6, 137.7, 138.2, 148.9, 168.6, 191.3. IR (KBr) v/cm-1: 3321, 3019, 2969, 1698, 1575, 1116. Anal. Calcd. for C23H21BrN4O2: C, 59.36; H, 4.55; N, 12.04; found: C, 59.46; H, 4.50; N, 12.14.
3-(2-(2,4-Difluorophenyl)-2-oxoethyl)-1-(3-oxo-3-(quinolin-8-ylamino)propyl)-1H-imidazol-3-ium chloride (8k)
White powder, 76% yield, m.p. = 221–223 °C. 1H-NMR (500 MHz, DMSO-d6): δ = 3.34 (at, J = 6.5 Hz, 2H, H11), 4.64 (t, J = 6.5 Hz, 2H, H12), 5.90 (s, 2H, H1’), 7.35 (td, J = 8.5 Hz, 2H, 2xH5’), 7.61–7.56 (m, 2H, H7’, H6), 7.65 (dd, J = 4.5 Hz, J = 8.5 Hz,1H, H3,), 7.70–7.69 (m, 2H, H5, H17), 7.92 (bs, 1H, H17), 8.05 (q, J = 8.5 Hz, 2H, 2xH4’), 8.42 (add, J = 8.5 Hz, 1H, H4), 8.59 (d, J = 7.5 Hz, 1H, H7), 8.94 (add, J = 4.5 Hz, 1H, H2), 9.21 (s, 1H, H14), 10.38 (s, 1H, H9). 13C-NMR (125 MHz, DMSO-d6): δ = 36.2, 45.2, 57.8, 105.5, 113.1, 117.3, 119.1, 122.2, 122.2, 122.3, 124.0, 126.8, 127.9, 132.7, 134.2, 136.6, 137.8, 138.2, 148.9, 162.5, 165.9, 168.6, 187.9. IR (KBr) v/cm−1: 3319, 3006, 2974, 1715, 1685, 1540, 1221, 1123. Anal. Calcd. for C23H19ClF2N4O2: C, 60.47; H, 4.19; N, 12.26; found: C, 60.57; H, 4.09; N, 12.16.
3-(2-(4-Nitrophenyl)-2-oxoethyl)-1-(3-oxo-3-(quinolin-8-ylamino)propyl)-1H-benzo[d]imidazol-3-ium bromide (12a)
Yellowish powder, 76% yield, m.p. = 165–168 °C. 1H-NMR (500 MHz, DMSO-d6): δ = 3.44 (at, J = 6.5 Hz, 2H, H11), 4.99 (t, J = 6.5 Hz, 2H, H12), 6.49 (s, 2H, H1’), 7.57 (t, J = 8.0 Hz, 1H, H6) 7.63 (dd, J = 4.5 Hz, J = 8.5 Hz,1H, H3), 7.70–7.67 (m, 2H, H5, H18), 7.75 (t, J = 8.5 Hz, 1H, H17), 8.09 (d, J = 8.5 Hz, 1H, H19), 8.25 (d, J = 8.5 Hz, 1H, H16), 8.34 (d, J = 8.5 Hz, 2H, 2xH4’), 8.40 (d, J = 8.5 Hz, 1H, H4), 8.46 (d, J = 8.5 Hz, 2H, 2xH5’), 8.57 (d, J = 8.0 Hz, 1H, H7), 8.91 (ad, J = 4.5 Hz, 1H, H2), 9.79 (s, 1H, H14), 10.38 (s, 1H, H9). 13C-NMR (125 MHz, DMSO-d6): δ = 35.2, 43.2, 53.7, 114.0, 114.1, 117.4, 122.2, 122.4, 124.1, 126.7, 126.8, 126.9, 127.9, 129.9, 130.7, 131.9, 134.2, 136.6, 138.3, 138.4, 143.9, 148.9, 150.6, 168.8, 190.6. IR (KBr) v/cm−1: 3334, 3013, 2981, 1712, 1675, 1547, 1339. Anal. Calcd. for C27H22BrN5O4: C, 57.87; H, 3.96; N, 12.50; found: C, 57.77; H, 3.91; N, 12.65.
3-(2-(4-Methoxyphenyl)-2-oxoethyl)-1-(3-oxo-3-(quinolin-8-ylamino)propyl)-1H-benzo[d]imidazol-3-ium bromide (12b)
White powder, 85% yield; m.p. = 220–221 °C. 1H-NMR (500 MHz, DMSO-d6): δ = 3.44 (at, J = 6.5 Hz, 2H, H11), 3.89 (s, 3H, p-OCH3), 4.97 (t, J = 6.5 Hz, 2H, H12), 6.36 (s, 2H, H1’), 7.17 (d, J = 9.0 Hz, 2H, 2xH5’), 7.58 (t, J = 8.0 Hz, 1H, H6), 7.69–7.63 (m, 3H, H3, H5, H18), 7.73 (t, J = 8.5 Hz, 1H, H17), 8.02 (d, J = 8.0 Hz, 1H, H19), 8.07 (d, J = 9.0 Hz, 2H, 2xH4’), 8.23 (d, J = 8.5 Hz, 1H, H16), 8.40 (add, J = 8.5 Hz, 1H, H4), 8.58 (d, J = 8.0 Hz, 1H, H7), 8.91 (add, J = 4.0 Hz, 1H, H2), 9.79 (s, 1H, H14), 10.38 (s, 1H, H9). 13C-NMR (125 MHz, DMSO-d6): δ = 35.2, 43.2, 52.8, 55.8, 113.9, 114.0, 114.3, 117.4, 122.2, 122.4, 126.6, 126.7, 126.8, 127.9, 130.7, 130.9, 131.9, 134.2, 136.6, 138.2, 144.0, 148.9, 164.2, 168.8, 189.4. IR (KBr) v/cm−1: 3247, 3002, 2986, 1675, 1601, 1548, 1243, 1165. Anal. Calcd. for C28H25BrN4O3: C, 61.66; H, 4.62; N, 10.27; found: C, 61.56; H, 4.60; N, 10.38.
3-(2-(4-Chlorophenyl)-2-oxoethyl)-1-(3-oxo-3-(quinolin-8-ylamino)propyl)-1H-benzo[d]imidazol-3-ium bromide (12c)
White powder, 86% yield; m.p. = 229–230 °C. 1H-NMR (500 MHz, DMSO-d6): δ = 3.44 (at, J = 6.0 Hz, 2H, H11), 4.98 (t, J = 6.0 Hz, 2H, H12), 6.42 (s, 2H, H1’), 7.57 (t, J = 8.0 Hz, 1H, H6), 7.63 (dd, J = 4.0 Hz, J = 8.0 Hz, 1H, H3), 7.69–7.66 (m, 2H, H15, H18), 7.75–7.72 (m, 3H, 2xH5’, H17), 8.05 (d, J = 8.5 Hz, 1H, H19), 8.12 (d, J = 8.5 Hz, 2H, 2xH4’), 8.24 (d, J = 8.5 Hz, 1H, H16), 8.40 (d, J = 8.0 Hz, 1H, H4), 8.57 (d, J = 8.0 Hz, 1H, H7), 8.91 (ad, J = 4.0 Hz, 1H, H2), 9.79 (s, 1H, H14), 10.38 (s, 1H, H9). 13C-NMR (125 MHz, DMSO-d6): δ = 35.2, 43.2, 53.3, 114.0, 117.4, 122.2, 122.4, 126.6, 126.8, 126.8, 127.9, 129.2, 130.3, 130.7, 131.9, 132.5, 134.2, 136.6, 138.2, 139.4, 143.9, 148.9, 168.8, 190.4. IR (KBr) v/cm−1: 3217, 3009, 2912, 1687, 1535, 739. Anal. Calcd. for C27H22BrClN4O2: C, 58.98; H, 4.03; N, 10.19; found: C, 58.88; H, 4.08; N, 10.09.
3-(2-Oxo-2-(p-tolyl)ethyl)-1-(3-oxo-3-(quinolin-8-ylamino)propyl)-1H-benzo[d]imidazol-3-ium bromide (12d)
White powder, 82% yield; m.p. = 225–227 °C. 1H-NMR (500 MHz, DMSO-d6): δ = 2.43 (s, 3H, p-CH3), 3.43 (at, J = 6.5 Hz, 2H, H11), 4.97 (t, J = 6.5 Hz, 2H, H12), 6.39 (s, 2H, H1’), 7.46 (d, J = 8.0 Hz, 2H, 2xH5’), 7.57 (t, J = 8.0 Hz, 1H, H6), 7.63 (dd, J = 4.0 Hz, J = 8.0 Hz, 1H, H3), 7.69–7.65 (m, 2H, H15, H18), 7.73 (t, J = 8.0 Hz, 1H, H17), 8.03–7.99 (m, 3H, 2xH4’, H19), 8.23 (d, J = 8.0 Hz, 1H, H16), 8.40 (d, J = 8.0 Hz, 1H, H4), 8.57 (d, J = 8.0 Hz, 1H, H7), 8.91 (ad, J = 4.0 Hz, 1H, H2), 9.79 (s, 1H, H14), 10.38 (s, 1H, H9). 13C-NMR (125 MHz, DMSO-d6): δ = 21.4, 35.2, 43.2, 53.3, 113.9, 114.0, 117.4, 122.2, 122.4, 126.6, 126.7, 126.8, 127.9, 128.5, 129.6, 130.7, 131.3, 131.9, 134.3, 136.6, 138.3, 144.0, 145.3, 148.9, 168.8, 190.7. IR (KBr) v/cm−1: 3284, 3035, 2971, 1696, 1612, 1574, 1213. Anal. Calcd. for C28H25BrN4O2: C, 63.52; H, 4.76; N, 10.58; found: C, 63.42; H, 4.66; N, 10.78.
3-(2-(4-Cyanophenyl)-2-oxoethyl)-1-(3-oxo-3-(quinolin-8-ylamino)propyl)-1H-benzo[d]imidazol-3-ium bromide (12e)
White powder, 90% yield; m.p. = 213–215 °C. 1H-NMR (500 MHz, DMSO-d6): δ = 3.46 (t, J = 6.5 Hz, 2H, H11), 4.99 (t, J = 6.5 Hz, 2H, H12), 6.47 (s, 2H, H1’), 7.57 (t, J = 7.5 Hz, 1H, H6), 7.63 (dd, J = 8.0 Hz, J = 4.0 Hz, 1H, H3), 7.69–7.66 (m, 2H, H15, H18), 7.75 (t, J = 7.5 Hz, 1H, H17), 8.09 (d, J = 8.0 Hz, 1H, H19), 8.16 (d, J = 8.5 Hz, 2H, 2xH5’), 8.26–8.24 (m, 2xH4’, 3H, H16), 8.40 (add, J = 8.0 Hz, 1H, H4), 8.58 (d, J = 7.5 Hz, 1H, H7), 8.92 (add, J = 4.0 Hz, 1H:, H2), 9.78 (s, 1H, H14), 10.38 (s, 1H, H9). 13C-NMR (125 MHz, DMSO-d6): δ = 35.2, 43.2, 53.5, 114.0, 116.1, 117.3, 118.0, 122.1, 122.3, 126.6, 126.7, 126.8, 127.8, 129.0, 130.7, 131.8, 133.0, 134.2, 136.6, 137.0, 138.2, 143.9, 148.9, 168.8, 190.8. IR (KBr) v/cm−1: 3312, 3005, 2943, 2230, 1716, 1535, 1227, 1073. Anal. Calcd. for C28H22BrN5O2: C, 62.23; H, 4.10; N, 12.96; found: C, 62.13; H, 4.15; N, 12.90.
3-(2-(4-Fluorophenyl)-2-oxoethyl)-1-(3-oxo-3-(quinolin-8-ylamino)propyl)-1H-benzo[d]imidazol-3-ium bromide (12f)
White powder, 81% yield; m.p. = 230–233 °C. 1H-NMR (500 MHz, DMSO-d6): δ = 3.46 (bs, 2H, H11), 4.99 (bs, 2H, H12), 6.44 (s, 2H, H1’), 7.74–7.51 (m, 7H, H3, H5, H6, 2xH5’, H17, H18), 8.07 (d, J = 7.5 Hz, 1H, H19), 8.25–8.20 (m, 3H, 2xH4’, H16), 8.41 (ad, J = 7.5 Hz, 1H, H4,), 8.58 (ad, 1H, J = 6.0 Hz, H7), 8.91 (abs, 1H, H2), 9.81 (s, 1H, H14), 10.39 (s, 1H, H9). 13C-NMR (125 MHz, DMSO-d6): δ = 35.2, 43.2, 53.2, 114.0, 116.2, 117.3, 122.1, 122.3, 126.5, 126.7, 126.8, 127.8, 130.5, 130.7, 131.6, 131.9, 134.2, 136.5, 138.2, 143.9, 148.9, 165.7, 168.7, 189.9. IR (KBr) v/cm−1: 3326, 3007, 2982, 1715, 1686, 1537, 1235, 1082. Anal. Calcd. for C27H22BrFN4O2: C, 60.80; H, 4.16; N, 10.50; found: C, 60.90; H, 4.11; N, 10.40.
3-(2-(4-Bromophenyl)-2-oxoethyl)-1-(3-oxo-3-(quinolin-8-ylamino)propyl)-1H-benzo[d]imidazol-3-ium bromide (12g)
White powder, 83% yield; m.p. = 213–215 °C. 1H-NMR (500 MHz, DMSO-d6): δ = 3.45 (at, J = 6.0 Hz, 2H, H11), 4.98 (at, J = 6.5 Hz, 2H, H12), 6.41 (s, 2H, H1’), 7.57 (t, J = 8.0 Hz, 1H, H6), 7.63 (dd, J = 4.0 Hz, J = 8.0 Hz, 1H, H3), 7.69–7.66 (m, 2H, H5, H18), 7.74 (t, J = 8.0 Hz, 1H, H17), 7.89 (d, J = 8.5 Hz, 2H, 2xH5’), 8.07–8.02 (d, 3H, 2xH4’, H19), 8.24 (d, J = 8.0 Hz, 1H, H16), 8.41 (add, J = 8.0 Hz, 1H, H4), 8.58 (d, J = 8.0 Hz, 1H, H7), 8.92 (add, J = 4.0 Hz, 1H, H2), 9.78 (s, 1H, H14), 10.38 (s, 1H, H9). 13C-NMR (125 MHz, DMSO-d6): δ = 35.1, 43.2, 53.2, 114.0, 117.3, 122.1, 122.3, 126.0, 126.7, 126.8, 127.8, 128.7, 130.3, 130.7, 131.8, 130.1, 132.8, 134.2, 136.6, 138.2, 143.9, 148.9, 168.7, 190.6. IR (KBr) v/cm−1: 3300, 3020, 2970, 1732, 1700, 1530, 1229, 1170, 670. Anal. Calcd. for C27H22Br2N4O2: C, 54.57; H, 3.73; N, 9.43; found: C, 54.47; H, 3.63; N, 9.63.
3-(2-([1,1′-Biphenyl]-4-yl)-2-oxoethyl)-1-(3-oxo-3-(quinolin-8-ylamino)propyl)-1H-benzo[d]imidazol-3-ium bromide (12h)
White powder, 81% yield; m.p. = 201–203 °C. 1H-NMR (500 MHz, DMSO-d6): δ = 3.47 (at, J = 6.0 Hz, 2H, H11), 5.00 (at, J = 6.0 Hz, 2H, H12), 6.47 (s, 2H, H1’), 7.47 (at, J = 7.5 Hz, 1H, H10’), 7.59–7.53 (m, 3H, H6, 2xH9’), 7.64 (dd, J = 4.5 Hz, J = 8.5 Hz, 1H, H3), 7.70–7.67 (m, 2H, H15, H18), 7.75 (t, J = 8.0 Hz, 1H, H17), 7.82 (d, J = 7.5 Hz, 2H, 2xH8’), 7.98 (d, J = 8.5 Hz, 2H, 2xH5’), 8.07 (d, J = 8.0 Hz,1H, H19), 8.19 (d, J = 8.5 Hz, 2H, 2xH4’), 8.25 (d, J = 8.0 Hz, 1H, H16), 8.41 (add, J = 8.5 Hz, 1H, H4), 8.59 (d, J = 7.5 Hz, 1H, H7), 8.92 (add, J = 4.5 Hz, 1H, H2) 9.82 (s, 1H, H14), 10.40 (s, 1H, H9). 13C-NMR (125 MHz, DMSO-d6): δ = 35.2, 43.2, 53.2, 114.0, 117.4, 122.1, 122.3, 126.6, 126.7, 126.8, 127.1, 127.9, 128.7, 129.1, 129.2, 130.7, 131.9, 132.5, 134.2, 136.6, 138.2, 138.5, 143.9, 145.7, 148.9, 168.8, 190.7. IR (KBr) v/cm−1: 3294, 3035, 2971, 1689, 1674, 1573, 1212. Anal. Calcd. for C33H27BrN4O2: C, 67.01; H, 4.60; N, 9.47; found: C, 67.11; H, 4.55; N, 9.37.
3-(2-Oxo-2-(4-(trifluoromethyl)phenyl)ethyl)-1-(3-oxo-3-(quinolin-8-ylamino)propyl)-1H-benzo[d]imidazol-3-ium bromide (12i)
White powder, 74% yield; m.p. = 217–218 °C. 1H-NMR (500 MHz, DMSO-d6): δ = 3.46 (at, J = 6.0 Hz, 2H, H11), 5.00 (at, J = 6.5 Hz, 2H, H12), 6.49 (s, 2H, H1’), 7.57 (t, J = 8.0 Hz, 1H, H6), 7.63 (dd, J = 4.0 Hz, J = 8.0 Hz, 1H, H3), 7.70–7.67 (m, 2H, H15, H18), 7.75 (t, J = 8.0 Hz, 1H, H17), 8.10–8.04 (m, 3H, 2xH5’, H19), 8.25 (d, J = 8.0 Hz, 1H, H16), 8.30 (d, J = 8.0 Hz, 2H, 2xH4’), 8.41 (add, J = 8.0 Hz, 1H, H4,), 8.58 (d, J = 8.0 Hz, 1H, H7), 8.92 (add, J = 4.0 Hz, 1H, H2), 9.80 (s, 1H, H14), 10.39 (s, 1H, H9). 13C-NMR (125 MHz, DMSO-d6): δ = 35.2, 43.2, 53.5, 114.0, 117.3, 122.1, 122.3, 123.6, 126.0, 126.6, 126.7, 126.8, 127.8, 129.3, 130.7, 131.8, 133.5, 134.2, 136.6, 137.0, 138.2, 143.9, 148.9, 168.7, 190.8. IR (KBr) v/cm−1: 3342, 3019, 2975, 1720, 1643, 1523, 1241, 1117. Anal. Calcd. for C28H22BrF3N4O2: C, 57.65; H, 3.80; N, 9.60; found: C, 57.55; H, 3.85; N, 9.50.
3-(2-Oxo-2-phenylethyl)-1-(3-oxo-3-(quinolin-8-ylamino)propyl)-1H-benzo[d]imidazol-3-ium bromide (12j)
White powder, 80% yield; m.p. = 196–198 °C. 1H-NMR (500 MHz, DMSO-d6): δ = 3.46 (at, J = 6.0 Hz, 2H, H11), 4.98 (at, J = 6.0 Hz, 2H, H12), 6.42 (s, 2H, H1’), 7.57 (t, J = 8.0 Hz, 1H, H6), 7.69–7.62 (m, 5H, H3, H5, 2xH5’, H6’), 7.74 (t, J = 8.0 Hz, 1H, H17), 7.79 (t, J = 8.0 Hz, 1H, H18), 8.05 (d, J = 8.0 Hz, 1H, H19), 8.11 (d, J = 7.5 Hz, 1H, 2xH4’), 8.24 (d, J = 8.0 Hz, 1H, H16), 8.41 (add, J = 8.0 Hz, 1H, H4,), 8.58 (d, J = 7.5 Hz, 1H, H7), 8.92 (add, J = 4.0 Hz, 1H, H2), 9.78 (s, 1H, H14), 10.39 (s, 1H, H9). 13C-NMR (125 MHz, DMSO-d6): δ = 35.1, 43.2, 53.2, 114.0, 117.3, 122.1, 122.3, 126.5, 126.7, 126.8, 127.8, 128.4, 129.0, 130.7, 131.9, 133.7, 134.2, 134.5, 136.6, 138.2, 143.9, 148.9, 168.7, 191.1. IR (KBr) v/cm−1: 3273, 3031, 2976, 1698, 1684, 1589, 1207. Anal. Calcd. for C27H23BrN4O2: C, 62.92; H, 4.50; N, 10.87; found: C, 62.82; H, 4.45; N, 10.97.
3-(2-(2,4-Difluorophenyl)-2-oxoethyl)-1-(3-oxo-3-(quinolin-8-ylamino)propyl)-1H-benzo[d]imidazol-3-ium chloride (12k)
White powder, 70% yield; m.p. = 203–205 °C. 1H-NMR (500 MHz, DMSO-d6): δ = 3.44 (at, J = 6.5 Hz, 2H, H11), 4.98 (t, J = 6.5 Hz, 2H, H12), 6.24 (s, 2H, H1’), 7.36 (td, J = 8.5 Hz, 1H, H5’), 7.57 (t, J = 7.5 Hz, 1H, H6), 7.64–7.60 (m, 2H, H3, H7’), 7.69–7.65 (m, 2H, H5, H18), 7.73 (t, J = 8.0 Hz,1H, H17), 8.09–8.05 (m, 2H, H4’, H19), 8.24 (d, J = 8.0 Hz, 1H, H16), 8.41 (add, J = 8.0 Hz, 1H, H4), 8.57 (d, J = 7.5 Hz, 1H, H7), 8.92 (add, J = 4.0 Hz, 1H, H2), 9.84 (s, 1H, H14), 10.39 (s, 1H, H9). 13C-NMR (125 MHz, DMSO-d6): δ = 35.2, 43.1, 55.8, 105.5, 113.0, 113.9, 114.2, 117.5, 119.3, 122.1, 122.4, 126.5, 126.7, 126.8, 127.9, 130.6, 131.9, 132.7, 134.2, 136.6, 138.2, 143.9, 148.9, 162.8, 166.0, 168.7, 187.7. IR (KBr) v/cm−1: 3332, 3006, 2973, 1698, 1680, 1575, 1397, 1231. Anal. Calcd. for C27H21ClF2N4O2: C, 63.97; H, 4.18; N, 11.05; found: C, 63.87; H, 4.13; N, 11.25.
Cell proliferation assay
The in vitro biological tests were performed to the National Cancer Institute (NCI, USA), under the Developmental Therapeutics Program (DTP). The DTP screens include the NCI 60 cell line screen and, as appropriate, the hollow fiber assay and relevant human tumor xenograft and rodent tumor models.
The operation of this screen utilizes 60 different human tumor cell lines, representing leukemia, melanoma and cancers of the lung, colon, brain, ovary, breast, prostate, and kidney. The aim is to prioritize for further evaluation26,38, synthetic compounds or natural product samples showing selective growth inhibition or cell killing of particular tumor cell lines. This screen is unique in that the complexity of a 60 cell line dose response produced by a given compound results in a biological response pattern which can be utilized in pattern recognition algorithms via COMPARE program (See: http://dtp.nci.nih.gov/docs/compare/compare.html).
The screening is beginning with the evaluation of all compounds against the 60 cell lines at a single dose of 10–5 M. The output from the single dose screen is reported as a mean graph and is available for analysis by the COMPARE program.
The standard NCI/DTP methodology of the in vitro cancer screen
See: https://dtp.cancer.gov/discovery_development/nci-60/methodology.html.
The human tumor cell lines of the cancer screening panel are grown in RPMI 1640 medium containing 5% fetal bovine serum and 2 mM l-glutamine. For a typical screening experiment, cells are inoculated into 96 well microtiter plates in 100 µL at plating densities ranging from 5000 to 40,000 cells/well depending on the doubling time of individual cell lines. After cell inoculation, the microtiter plates are incubated at 37 °C, 5% CO2, 95% air and 100% relative humidity for 24 h prior to addition of experimental drugs.
After 24 h, two plates of each cell line are fixed in situ with TCA, to represent a measurement of the cell population for each cell line at the time of drug addition (Tz). Experimental drugs are solubilized in dimethyl sulfoxide at 400-fold the desired final maximum test concentration and stored frozen prior to use. At the time of drug addition, an aliquot of frozen concentrate is thawed and diluted to twice the desired final maximum test concentration with complete medium containing 50 µg/ml gentamicin. Additional four, tenfold or ½ log serial dilutions are made to provide a total of five drug concentrations plus control. Aliquots of 100 µL of these different drug dilutions are added to the appropriate microtiter wells already containing 100 µL of medium, resulting in the required final drug concentrations.
Following drug addition, the plates are incubated for an additional 48 h at 37 °C, 5% CO2, 95% air, and 100% relative humidity. For adherent cells, the assay is terminated by the addition of cold TCA. Cells are fixed in situ by the gentle addition of 50 µL of cold 50% (w/v) TCA (final concentration, 10% TCA) and incubated for 60 min at 4 °C. The supernatant is discarded, and the plates are washed five times with tap water and air dried. Sulforhodamine B (SRB) solution (100 µL) at 0.4% (w/v) in 1% acetic acid is added to each well, and plates are incubated for 10 min at room temperature. After staining, unbound dye is removed by washing five times with 1% acetic acid and the plates are air dried. Bound stain is subsequently solubilized with 10 mM trizma base, and the absorbance is read on an automated plate reader at a wavelength of 515 nm. For suspension cells, the methodology is the same except that the assay is terminated by fixing settled cells at the bottom of the wells by gently adding 50 µL of 80% TCA (final concentration, 16% TCA). Using the seven absorbance measurements [time zero, (Tz), control growth, (C), and test growth in the presence of drug at the five concentration levels (Ti)], the percentage growth is calculated at each of the drug concentrations levels. Percentage growth inhibition (PGI) is calculated as:
Three dose response parameters are calculated for each experimental agent. Growth inhibition of 50% (GI50) is calculated from [(Ti − Tz)/(C − Tz)] × 100 = 50, which is the drug concentration resulting in a 50% reduction in the net protein increase (as measured by SRB staining) in control cells during the drug incubation. The drug concentration resulting in total growth inhibition (TGI) is calculated from Ti = Tz. The LC50 (concentration of drug resulting in a 50% reduction in the measured protein at the end of the drug treatment as compared to that at the beginning) indicating a net loss of cells following treatment is calculated from [(Ti − Tz)/Tz] × 100 = − 50. Values are calculated for each of these three parameters if the level of activity is reached; however, if the effect is not reached or is exceeded, the value for that parameter is expressed as greater or less than the maximum or minimum concentration tested.
Antimicrobial assay
Disk-diffusion method
The in vitro antibacterial and antifungal activity of the hybrid quinoline—imidazole/benzimidazole compounds were determined by the Kirby-Bauer disk diffusion method41. The method is using Mueller Hinton agar medium for antibacterial assay and Sabouraud nutrient agar medium for antifungal assay. The in vitro antibacterial activity was evaluated against Gram-positive bacteria Staphylococcus aureus ATCC 25923 and Gram-negative Escherichia coli ATCC 25922 and the antifungal activity against fungus Candida albicans ATCC 1023123. Gentamicin was used as positive control (C+) for S. aureus and E. coli and Nystatin for C. albicans. The negative control (C−) consist of sterile filter paper disks (with no antimicrobial compounds) inoculated with DMSO 3%.
For inoculum preparation, reference microbial cultures of bacteria (Staphylococcus aureus ATCC 25923, Escherichia coli ATCC 25922) and fungi (Candida albicans ATCC 10231) were employed. Approximately 5 colonies from each type of culture were used to inoculate 10 mL of Mueller Hinton (MH) agar (for antibacterial tests) and Sabouraud agar (for antifungal tests). Using a Beckman Coulter DU 730 spectrophotometer (λ = 600 nm), the turbidity of the inoculum was adjusted to a 0.5 McFarland standard (1–2 × 108 CFU/mL for bacteria and 1–5 × 106 CFU/mL for Candida), and the inoculum was transferred, in a 1 mL volume, onto the surface of the growth media specific for bacteria (MH) and fungi (Sabouraud). Once the inoculum was absorbed, sterile paper disks of approximately 6 mm in diameter and impregnated with 10 µL of antibacterial compound (dissolved in DMSO 3%) were placed on the surface of the culture media; for all the tested compounds, the concentration used was 25 mg/mL. Following incubation at the optimal temperatures for bacteria and fungi, of 37 °C and 28 °C, respectively, for 24 h (bacteria) and 72 h (fungi), the diameters of the inhibition zones were measured using a ruler. The controls were prepared in the same growth conditions (i.e. C+: sterile filter paper disks impregnated with antibiotics inducing sensitivity in the organisms under investigation, namely Gentamicin 10 μg/mL for Staphylococcus aureus and Escherichia coli and Nystatin 100 μg/mL for Candida albicans, and C-: sterile filter paper disks with no antimicrobial compounds − 10 µL DMSO 3%).
Broth microdilution method for determining the minimum inhibitory concentration (MIC)
For the minimum inhibitory concentration (MIC) assay was used the 96-well microtiter plate (microdilution) technique43,44. The MIC was evaluated against Gram-positive bacteria Staphylococcus aureus ATCC 25923 and Gram-negative Escherichia coli ATCC 25922 and the antifungal activity against fungus Candida albicans ATCC 1023123,35. For each tested microorganism, a positive control C+ (containing 80 µL of MH growth medium and 10 µL of antimicrobial compound) and a negative one C− (containing 80 µL of MH growth medium and 10 µL of diluted microbial culture) were prepared. The redox dye, resazurin, was used as colorimetric indicator.
The working technique involves the use of a 96-well microtiter plate (microdilution). In each well of the plate, 80 µL of growth medium MH, 10 µL of microbial inoculum (Staphylococcus aureus ATCC 25923, Escherichia coli ATCC 25922, or Candida albicans ATCC 10231) were prepared in the same manner as in the diffusion test (i.e. by diluting the standardized microbial suspension adjusted to a 0.5 McFarland standard), and 100 µL of antimicrobial substance to be tested were transferred by pipetting, in different concentrations. To this purpose, double dilutions of the antimicrobial agent were made in the DMSO 3%, starting with the 25 µg/mL dilution (e.g. 12.5 µg/mL, 6.25 µg/mL, 3.12 µg/mL, 1.56 µg/mL, 0.78 µg/mL and so on). For each tested microorganism, a positive control C+ (containing 80 µL of MH growth medium and 10 µL of antimicrobial compound) and a negative one C− (containing 80 µL of MH growth medium and 10 µL of diluted microbial culture) were prepared. Following the incubation of the microplates at 37 °C for 24 h (for Staphylococcus aureus ATCC 25923 and Escherichia coli ATCC 25922) and at 28 °C for 72 h (for Candida albicans ATCC 10231), 10 µL of resazurin were added in each well. The samples were incubated once again at the temperature optimal for each microorganism for one hour. The colour of the indicator turned from purple to pink. Resazurin is a colorimetric indicator for cell viability widely applied for monitoring cell proliferation46. The redox dye, resazurin, enters the cytosol in the oxidized form (purple-blue) and is converted to the reduced form, resorufin (pink).
Data availability
All data generated or analysed during this study are included in this published article [and its supplementary information files].
References
WHO. Cancer. https://www.who.int/news-room/fact-sheets/detail/cancer. Accessed 5 Feb 2022.
Vitaku, E., Smith, D. T. & Njardarson, J. T. Analysis of the structural diversity, substitution patterns, and frequency of nitrogen heterocycles among US FDA approved pharmaceuticals. J. Med. Chem. 57, 10257–10274. https://doi.org/10.1021/jm501100b (2014).
Abbot, V. et al. Small hybrid heteroaromatics: Resourceful biological tools in cancer research. RSC Adv. 7, 28313–28349. https://doi.org/10.1039/C6RA24662A (2017).
Akhtar, J. et al. Structure-activity relationship (SAR) study and design strategies of nitrogen-containing heterocyclic moieties for their anticancer activities. Eur. J. Med. Chem. 125, 143–189. https://doi.org/10.1016/j.ejmech.2016.09.023 (2017).
Nepali, K. et al. Rational approaches, design strategies, structure activity relationship and mechanistic insights for anticancer hybrids. Eur. J. Med. Chem. 77, 422–487. https://doi.org/10.1016/j.ejmech.2014.03.018 (2014).
WHO. Global Strategy for Containment of Antimicrobial Resistance. https://www.who.int/drugresistance/WHO_Global_Strategy_English.pdf. Accessed 5 Feb 2022.
Frieri, M., Kumar, K. & Boutin, A. Antibiotic resistance. J. Infect. Public. Heal. 10, 369–378. https://doi.org/10.1016/j.jiph.2016.08.007 (2017).
Rodrigues, M. E., Silva, S., Azeredo, J. & Henriques, M. Novel strategies to fight Candida species infection. Crit. Rev. Microbiol. 42, 594–606. https://doi.org/10.3109/1040841X.2014.974500 (2016).
Matada, B. S., Pattanashettar, R. & Yernale, N. G. A comprehensive review on the biological interest of quinoline and its derivatives. Bioorg. Med. Chem. 32, 115973. https://doi.org/10.1016/j.bmc.2020.115973 (2021).
Kumari, L. S., Mazumder, A., Kumar, V. & Gupta, S. Synthesis and biological potentials of quinoline analogues: A review of literature. Mini Rev. Org. Chem. 16, 653–688. https://doi.org/10.2174/1570193X16666190213105146 (2019).
Akhtar, W. et al. Therapeutic evolution of benzimidazole derivatives in the last quinquennial period. Eur. J. Med. Chem. 126, 705–753. https://doi.org/10.1016/j.ejmech.2016.12.010 (2017).
Salahuddin, M. S. & Mazumder, A. Benzimidazoles: A biologically active compounds. Arab. J. Chem. 10, S157–S173. https://doi.org/10.1016/j.arabjc.2012.07.017 (2017).
Chopra, P. & Sahu, J. Biological significance of imidazole-based analogues in new drug development. Curr. Drug. Discov. Technol. 17, 574–584. https://doi.org/10.2174/1570163816666190320123340 (2020).
Lauria, A., La Monica, G., Bono, A. & Martorana, A. Quinoline anticancer agents active on DNA and DNA-interacting proteins: From classical to emerging therapeutic targets. Eur. J. Med. Chem. 220, 113555. https://doi.org/10.1016/j.ejmech.2021.113555 (2021).
Gao, F., Zhang, X., Wang, T. & Xiao, J. Quinolone hybrids and their anticancer activities: An overview. Eur. J. Med. Chem. 165, 59–79. https://doi.org/10.1016/j.ejmech.2019.01.017 (2019).
Zhang, J., Wang, S., Ba, Y. & Xu, Z. 1,2,4-Triazole-quinoline/quinolone hybrids as potential antibacterial agents. Eur. J. Med. Chem. 174, 1–8. https://doi.org/10.1016/j.ejmech.2019.04.033 (2019).
Hu, Y. Q. et al. Quinoline hybrids and their antiplasmodial and antimalarial activities. Eur. J. Med. Chem. 139, 22–47. https://doi.org/10.1016/j.ejmech.2017.07.061 (2017).
Kakkar, A. K. & Dahiya, N. Bedaquiline for the treatment of resistant tuberculosis: promises and pitfalls. Tuberculosis 94, 357–362. https://doi.org/10.1016/j.tube.2014.04.001 (2014).
Gaba, M. & Singh, S. Benzimidazole: An emerging scaffold for analgesic and anti- inflammatory agents. Eur. J. Med. Chem. 76, 494–505. https://doi.org/10.1016/j.ejmech.2014.01.030 (2014).
Fang, Y., Zhou, H., Gu, Q. & Xu, J. Synthesis and evaluation of tetrahydroisoquinoline-benzimidazole hybrids as multifunctional agents for the treatment of Alzheimer’s disease. Eur. J. Med. Chem. 167, 133–145. https://doi.org/10.1016/j.ejmech.2019.02.008 (2019).
Diaconu, D. et al. Ultrasound assisted synthesis of hybrid quinoline-imidazole derivatives: a green synthetic approach. RSC Adv. 11, 38297–38301. https://doi.org/10.1039/D1RA07484A (2021).
Antoci, V. et al. Bis-(imidazole/benzimidazole)-pyridine derivatives: Synthesis, structure and antimycobacterial activity: Part XII. Future Med. Chem. 12, 207–222. https://doi.org/10.4155/fmc-2019-0063 (2020).
Diaconu, D. et al. Hybrid quinoline-sulfonamide complexes (M2+) derivatives with antimicrobial activity. Molecules 25, 2946. https://doi.org/10.3390/molecules25122946 (2020).
Lungu, C. N. et al. Hybrid imidazole: Pyridine derivatives: computational approach to novel anticancer DNA intercalators. Curr. Med. Chem. 27, 154–169. https://doi.org/10.2174/0929867326666181220094229 (2020).
Al Matarneh, C. et al. Synthesis and antibacterial evaluation of new pyrrolo[3’,4’:3,4]pyrrolo[1,2-a]quinoline and pyrrolo[3’,4’:3,4] pyrrolo[1,2-a]isoquinoline derivatives. Stud. UBB Chem. 64(3), 67–80 (2019).
Mantu, D. et al. Hybrid imidazole (benzimidazole)/pyridine (quinoline) derivatives and evaluation of their anticancer and antimycobacterial activity. J. Enzyme Inhib. Med. Chem. 31, 96–103. https://doi.org/10.1080/14756366.2016.1190711 (2016).
Panda, P. & Chakroborty, S. Navigating the synthesis of quinoline hybrid molecules as promising anticancer agents. Chem. Sel. 5, 10187–10199. https://doi.org/10.1002/slct.202002790 (2020).
Raghavendra, N. M. et al. Dual or multi-targeting inhibitors: The next generation anticancer agents. Eur. J. Med. Chem. 143, 1277–1300. https://doi.org/10.1016/j.ejmech.2017.10.021 (2018).
Rana, A. et al. A review on pharmacophoric designs of antiproliferative agents. Med. Chem. Res. 24, 903–920. https://doi.org/10.1016/j.ejmech.2017.10.021 (2015).
Bansal, Y. & Silakari, O. Multifunctional compounds: Smart molecules for multifactorial diseases. Eur. J. Med. Chem. 76, 31–42. https://doi.org/10.1016/j.ejmech.2014.01.060 (2014).
Sangani, C. B. et al. Design, synthesis and molecular modeling of pyrazole-quinoline-pyridine hybrids as a new class of antimicrobial and anticancer agents. Eur. J. Med. Chem. 76, 549–557. https://doi.org/10.1016/j.ejmech.2014.01.018 (2014).
Bremner, J. B., Ambrus, J. I. & Samosorn, S. Dual action-based approaches to antibacterial agents. Curr. Med. Chem. 14, 1459–1477. https://doi.org/10.2174/092986707780831168 (2007).
Castagnolo, D. et al. Synthesis, biological evaluation and SAR study of novel pyrazole analogues as inhibitors of Mycobacterium tuberculosis. Bioorg. Med. Chem. 16, 8587–8591. https://doi.org/10.1016/j.bmc.2008.08.016 (2008).
Danac, R., Daniloaia, T., Antoci, V. & Mangalagiu, I. I. Design, synthesis and antimycobacterial activity of some new azaheterocycles: Phenanthroline with p-halogeno-benzoyl skeleton: Part V. Lett. Drug Des. Discov. 12, 14–19 (2015).
Antoci, V. et al. Benzoquinoline derivatives: A straightforward and efficient route to antibacterial and antifungal agents. Pharmaceuticals 14, 335. https://doi.org/10.3390/ph14040335 (2021).
Hu, H. et al. Discovery of novel c-nesenchymal-epithelia transition factor and histone deacetylase dual inhibitors. Eur. J. Med. Chem. 204, 112651. https://doi.org/10.1016/j.ejmech.2020.112651 (2020).
Macıas, B. et al. Synthesis and structural characterization of zinc complexes with sulfonamides containing 8-aminoquinoleine. Z. Anorg. Allg. Chem. 629, 255–260 (2003).
Shoemaker, H. R. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer 6, 813–823. https://doi.org/10.1038/nrc1951 (2006).
Skehan, P. et al. New colorimetric cytotoxicity assay for anticancer-drug screening. J. Natl. Cancer Inst. 82, 1107–1112. https://doi.org/10.1093/jnci/82.13.1107 (1990).
Boyd, R. B. The NCI in vitro anticancer drug discovery screen. In Anticancer Drug Development Guide: Cancer Drug Discovery and Development (ed. Teicher, B. A.) 23–42 (Humana Press, 1997). https://doi.org/10.1007/978-1-4615-8152-9_2.
Weinstein, M. P. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 11th Edition, CLSI document M07-A11. (Clinical and Laboratory Standards Institute, 2018). https://clsi.org/media/1928/m07ed11_sample.pdf. Accessed 10 Feb 2022.
Konaté, K. et al. Anti-bacterial activity against β-lactamase producing methicillin and ampicillin-resistants Staphylococcus aureus: Fractional inhibitory concentration index (FICI) determination. Ann. Clin. Microbiol. Antimicrob. 11, 18. https://doi.org/10.1186/1476-0711-11-18 (2012).
Kavanagh, A. Effects of microplate type and broth additives on microdilution MIC susceptibility assays. Antimicrob. Agents Ch. 63, e01760-e1818. https://doi.org/10.1128/AAC.01760-18 (2019).
Balouiri, M., Sadiki, M. & Ibnsouda, K. S. Methods for in vitro evaluating antimicrobial activity. J. Pharm. Anal. 6, 71–79. https://doi.org/10.1016/j.jpha.2015.11.005 (2016).
Thakur, D. & Sahani, K. In vitro antimicrobial activity and MIC of the extracellular ethyl acetate crude extract of endophytic fungi Fusarium sp. isolated from Tephrosia purpurea root. Int. J. Pharm. Pharm. Sci. 11, 48–53 (2019).
Osaka, I. & Hefty, P. S. Simple resazurin-based microplate assay for measuring chlamydia infections. Antimicrob. Agents Chem. 57, 2838–2840. https://doi.org/10.1128/AAC.00056-13 (2013).
Acknowledgements
Authors are thankful for financial support to CNCS-UEFISCDI, project number PN-III-P4-IDPCE-2020-0371. The authors are also thankful to Romanian Ministry of Research, Innovation and Digitization, within Program 1—Development of the national RD system, Subprogram 1.2—Institutional Performance—RDI excellence funding projects, Contract no.11PFE/30.12.2021 and project POC/448/1/1 Research Centre with Integrated Techniques for Atmospheric Aerosol Investigation in Romania-RECENT AIR (grant agreement MySMIS no. 127324) for infrastructure used. The authors gratefully acknowledge to Prof. Simona Dunca for her help in antimicrobial assay, NCI (National Cancer Institute) for anticancer assay and CERNESIM, for NMR experiments.
Author information
Authors and Affiliations
Contributions
Design, conception and writing were performed by D.A.-M. and I.I.M. Synthesis, structure elucidation and biological data analysis were performed by all authors, which also reviewed and approved the final version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Diaconu, D., Antoci, V., Mangalagiu, V. et al. Quinoline–imidazole/benzimidazole derivatives as dual-/multi-targeting hybrids inhibitors with anticancer and antimicrobial activity. Sci Rep 12, 16988 (2022). https://doi.org/10.1038/s41598-022-21435-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-21435-6
- Springer Nature Limited
This article is cited by
-
Synthesis of Trifluoromethyl Benzimidazoles with Potential Anti-Microbial Activities
Iranian Journal of Science (2024)
-
Recent pharmacological insights about imidazole hybrids: a comprehensive review
Medicinal Chemistry Research (2024)
-
Synthesis and in-vitro anti-proliferative with antimicrobial activity of new coumarin containing heterocycles hybrids
Scientific Reports (2023)