Abstract
Improving predictive models for intensive care unit (ICU) inpatients requires a new strategy that periodically includes the latest clinical data and can be updated to reflect local characteristics. We extracted data from all adult patients admitted to the ICUs of two university hospitals with different characteristics from 2006 to 2020, and a total of 85,146 patients were included in this study. Machine learning algorithms were trained to predict in-hospital mortality. The predictive performance of conventional scoring models and machine learning algorithms was assessed by the area under the receiver operating characteristic curve (AUROC). The conventional scoring models had various predictive powers, with the SAPS III (AUROC 0.773 [0.766–0.779] for hospital S) and APACHE III (AUROC 0.803 [0.795–0.810] for hospital G) showing the highest AUROC among them. The best performing machine learning models achieved an AUROC of 0.977 (0.973–0.980) in hospital S and 0.955 (0.950–0.961) in hospital G. The use of ML models in conjunction with conventional scoring systems can provide more useful information for predicting the prognosis of critically ill patients. In this study, we suggest that the predictive model can be made more robust by training with the individual data of each hospital.
Similar content being viewed by others
Introduction
Intensive care unit (ICU) is a special setting in which patients have a higher baseline severity at the time of admission and hospital-associated infections occur more frequently than in general wards1,2. Due to the high mortality rate of ICU patients, several scoring systems have been used to predict patient’s prognosis3,4. Conventional scoring systems such as the Assessment and Chronic Health Evaluation (APACHE) II and III, the Simplified Acute Physiology Score (SAPS) II and III, the Mortality Probability Model (MPM0) II and III, and the Sequential Oran Failure Assessment (SOFA) are moderately accurate in predicting individual mortality1,3,4,5,6,7,8. The Pitt bacteremia score has also been shown to predict the prognosis even in patients with nonbacterial infections9.
These scoring systems have the advantage of standardizing studies and allowing the comparison of quality of care between ICUs. However, the prognostic predictive power of conventional models may vary depending on the characteristics of the hospitals in which they are to be applied, such as the demographic composition of inpatients, underlying comorbidities, and the percentage of surgical or medical admissions10,11,12. In addition, since conventional scoring systems were established and validated at specific points in time, predictive performance can deteriorate over time due to temporal and social changes, including an extension of life expectancy, improvements in the public health environment, and the emergence of new diseases10,11,12,13.
To improve the predictive model for ICU inpatients, it is necessary to prepare a new strategy that can be easily updated periodically to include the latest clinical data and to reflect the local features of each hospital14. The growth of automated electronic health records (EHRs) has provided an opportunity to extract a large amount of clinical information for improving the performance of established models. Additionally, machine learning (ML) techniques are being implemented in various ways for diagnosis and prognosis prediction in the field of clinical medicine including critical care medicine14,15,16,17,18. Comprehensive data analysis through ML can leverage the vast number of variables already inputted into EHRs to prevent overfitting and facilitate the development of continuously updatable prediction models14.
The purpose of this study was to show the importance of an updatable prognostic prediction model that can reflect the characteristics of individual centers. We compared the predictive models of ICU inpatients with conventional scoring systems and ML algorithms. Furthermore, we also evaluated the conventional models by year and hospital and, through ML algorithms, investigated variables that mutually affected the prognosis of patients admitted to the ICUs.
Methods
Study population and data collection
To determine a ML and statistical analysis approach based on local EHR data for mortality prediction, we extracted data from all adult patients (age ≥ 18 years) admitted to medical, surgical, and cardiovascular ICUs from 2006 to 2020 using the Severance Clinical Research Analysis Portal [EHR data collection program with information since 2006 of two university hospitals in South Korea. As of October 2021, this portal contains the data of over four million patients in Severance Hospital (hospital S) and over two million patients in Gangnam Severance Hospital (hospital G) and is continuously updated in real-time with only a short delay]. To compare the differences in the modeling fit according to hospital characteristics, data were collected from two different university hospitals [hospital S and hospital G, tertiary university hospitals with more than 2000 (including more than 130 adult ICU beds) and 800 beds (including more than 40 adult ICU beds), respectively] during the study period.
Patient-level data were collected, including demographics, underlying comorbidities, date of first ICU admission, and date of death. To obtain the worst values within the first 24 h of each ICU stay, both maximum and minimum values of vital signs and laboratory test results were extracted. The use of mechanical ventilation, vasopressors, and antibiotics within the first 24 h of admission to the ICUs was investigated, and the lowest Glasgow Coma Scale and total urine output per day were also extracted. The site of infection were defined according to the criteria proposed by the Centers for Disease Control and Prevention and a previous study19,20. Predictive models were developed by selecting in-hospital mortality as the outcome of interest.
All processes of this study were in accordance with the ethical standards of the institutional and national research committee and with the 1964 Helsinki declaration and comparable ethical standards. The study was approved by the Institutional Review Board (approval no.: 2021–0575–001) of Yonsei University Health System (Seoul, Republic of Korea), which waived the requirement for written informed consent because of the retrospective nature of this study.
Model development
Before the modeling, the type of admission, sex, type of admitted ICU, first site of bacterial infection, antibiotic use, and underlying comorbidities were treated as categorical values. In addition, continuous variables were standardized, and missing values were imputed using the median. A total of 70 features were included in the machine learning models.
The cohort C dataset included a random sampling of cases from hospital S with the same size as cases in hospital G, followed by a pooling of cases from both hospitals. Cohort S and cohort G included datasets from hospital S and hospital G patients, respectively (Fig. 1). The dataset was randomly split for all models. Eighty percent of the cases were used to train the scoring systems, and 20% were left as an internal test set for model evaluation. In addition, the model selected by training with cohort S was externally validated with cohort G and vice versa.
Candidate models were trained using K-nearest neighbor (KNN), decision tree (DT), random forest (RF), extreme gradient boosting (XGBoost), light gradient boosting (LightGBM), support vector machine (SVM) and artificial neural network (ANN) algorithms. ANN models were constructed with three layers with one input layer, two hidden layers (with 128 nodes applied), and one output layer. Models were trained to use the Root Mean Square Propagation algorithm to optimize the cross-entropy loss. For each model, hyperparameter tuning was randomly performed through grid search and fivefold cross-validation to find the optimal model while preventing overfitting. Each model with the highest area under the receiver operator characteristic curve (AUROC) and the 95% confidence intervals (CIs) were generated using bootstrapping14,21. We also used the F1 score (the harmonic mean of precision and recall), accuracy, and area under the precision-recall curve (AUPRC) as our secondary performance metrics for model comparison, as they are more informative for predicting in-hospital mortality15,22,23. To explain the model, we visualized feature importance rankings as feature weights, mean gain, and coverage for the XGBoost models and Shapley additive explanation (SHAP) summary dot plots for the LightGBM models24. We also presented calibration plots to illustrate each model’s calibration of consistency between observed and estimated in-hospital mortality.
ML models were analyzed using Python programming software version 3.7.12 (Python software foundation, http://www.python.org/) with the scikit-learn version 0.22.2 and TensorFlow version 2.6.0 libraries.
Statistical analysis
All variables were assessed by the Shapiro–Wilk test to evaluate the normality of the distributions, and admission variables are described using numbers and percentages for categorical variables, medians, and interquartile ranges (IQRs) for continuous variables. Differences were tested with Fisher's exact test for qualitative data and with the Mann–Whitney U test for quantitative data. All reported p values were 2-sided, and statistical significance was assumed if P < 0.05. Statistical analyses were performed using R statistical software version 4.1 (R Studio, Inc., http://www.r-project.org/).
Ethical approval
The study was approved by the Institutional Review Board (approval no.: 2021–0575–001) of Yonsei University Health System (Seoul, Republic of Korea), which waived the requirement for written informed consent because of the retrospective nature of this study.
Results
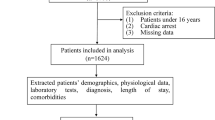
Figure 1 is a flowchart showing the dataset selection process for modeling. During the study period, 109,708 patients were admitted to the medical, surgical, and cardiovascular ICUs of hospital S and hospital G. After excluding multiple admissions except for the first admission, patients without demographic data, and patients missing more than 20% of the variables, a total of 85,146 patients were included in the study.
The clinical and demographic characteristics are described in Table 1. The median age of the patients was 67 years (IQR, 57–74 years) in hospital S and 65 years (IQR, 53–75 years) in hospital G. There were statistically significant differences in all variables between patients with and without in-hospital mortality at hospital S. Similarly, in hospital G, all variables were significantly different between the two groups except for sex, history of cerebrovascular disease, and history of diabetes mellitus. In both cohorts, patients with in-hospital death were older, were more likely to be admitted for medical than surgical reasons, had higher scores on the conventional prediction model and the Charlson comorbidity index than patients without in-hospital death. The in-hospital mortality rates of hospital S and hospital G were 11.8% and 17.4%, respectively, which were significantly different (P < 0.001). Additionally, the scores of all conventional prediction models except MPM0 were significantly higher in hospital G than in hospital S (P < 0.001).
There was a significant difference in the composition of patients admitted to the ICUs of the two cohorts. Hospital G had a higher rate of admission due to surgical factors than hospital S (51.8% vs. 35.8%, P < 0.001). The rate of cardiac arrest within 24 h of ICU admission was significantly higher in hospital G than in hospital S (5.2% vs. 1.3%; P < 0.001). In both hospitals, positive bacterial culture results were associated with a higher mortality rate than culture-negative or nonmicrobiological test prescriptions, but the bacterial culture positive rates in each cohort showed a statistically significant difference (3.4% in hospital S vs. 11.3% in hospital G; P < 0.001).
The proportions of departments at the time of ICU admission in each cohort also differed significantly (Table S1; P < 0.001). In hospital S, Departments of Cardiology, Neurology, and Cardiac Surgery had the highest proportions of inpatients (53.06%, 18.58%, and 17.38%, respectively), while in hospital G, the values for the Departments of Cardiology, Neurosurgery, and Thoracic Surgery were 25.33%, 20.5%, and 9.3%, respectively.
Table 2 shows that the conventional scoring systems had various predictive powers for each hospital and different study periods. For each study period, the highest AUROC values were 0.757 (95% CI, 0.744–0.769; SAPS II in 2005–2010), 0.777 (95% CI, 0.767–0.788; SAPS III in 2011–2015), and 0.808 (95% CI, 0.798–0.817; APACHE II in 2016–2020) in hospital S and 0.805 (95% CI, 0.790–0.819; APACHE III in 2005–2010), 0.789 (95% CI, 0.776–0.802; APACHE III and 95% CI, 0.773–0.799; SAPS III in 2011–2015), 0.831 (95% CI, 0.820–0.842; SAPS III in 2016–2020) in hospital G. During the entire study period, SAPS III showed the highest AUROC value (0.773 [95% CI, 0.766–0.779]) in hospital S, followed by SAPS II (0.766 [95% CI, 0.759–0.772]) and APACHE III (0.755 [95% CI, 0.747–0.762]). However, during certain periods, such as 2016–2020, APACHE II had the highest AUROC. Similarly, in hospital G, the best predictive model differed by study period. This performance was calculated by applying an optimal cutoff value (the point with the highest Youden’s index25) with the highest AUROC, and we found that these cutoff values increased over time for most models.
ML model
Table 3 summarizes the performance of ML algorithms for each dataset. For cohort C to equally reflect the features of the two hospitals, the cases from the hospital S dataset were randomly selected and combined with the same size as the cases from the hospital G dataset. For all variables, there were no significant differences between the randomly selected dataset and the full data of hospital S (Table S2). For cohort S and cohort G, ML was performed using the datasets of hospital S and hospital G, respectively. For cohort C, the XGBoost and LightGBM classifiers showed the highest AUROC value of 0.961 (95% CI, 0.957–0.965 for both), and the LightGBM classifier had the highest F1 score of 0.765. The classifiers that performed best with cohort S and cohort G were different. For cohort S, the XGBoost classifier achieved the highest values in both the F1 score (0.840) and AUROC (0.977; 95% CI, 0.973–0.980), and for cohort G, the LightGBM classifier showed both the highest F1 score (0.762) and the highest AUROC (0.955; 95% CI, 0.950–0.961). All ML algorithms (except for KNN with cohort C and cohort G) showed outstanding discriminant performance with an AUROC of 0.9 or higher26 and had superior performance to the conventional scoring systems in terms of the AUROC, accuracy, and F1 score. Figure 2 present the AUROCs, AUPRCs, and calibration plots of cohort C (Fig. 2a–c), cohort S (Fig. 2d–f), and cohort G (Fig. 2g–i) for in-hospital mortality. In all three models, the LightGBM classifier achieved the best AUPRC.
Comparison of machine learning-based in-hospital mortality prediction models. AUROC area under the receiver operating characteristic curve, AUPRC area under the precision-recall curve, KNN K-nearest neighbor, DT decision tree, RF random forest, XGBoost eXtreme gradient boosting, LightGBM light gradient boosting, SVM support vector machine. The calibration plots show the agreement of between predicted probability and observed in-hospital mortality. The black line at 45 degrees indicates perfect calibration where the predicted and observed probabilities are equal.
The results of external validation are described in Table S3. We validated the ML model obtained from cohort S with the dataset of cohort G and vice versa. The AUROCs of the models deteriorated to 0.654 (for the XGBoost classifier; 95% CI, 0.648–0.662) and 0.586 (for the LightGBM classifier; 95% CI, 0.581–0.591).
Critical variables with feature importance plots and SHAP values for predicting in-hospital mortality
The top predictors for in-hospital mortality obtained in each cohort are described in Fig. 3 (a–d, and d-f represent cohort C, cohort S, and cohort G, respectively). Figure 3a,c,e show the most important features calculated using the XGBoost models for in-hospital mortality. In cohort C, maximum serum glucose and positive blood culture results; in cohort S, patient age, positive blood culture results, and history of antibiotic use; and in cohort G, patient age and ventilator application were the top predictors for in-hospital mortality. SHAP analysis of the LightGBM models was also performed to investigate which individual variables impact in-hospital mortality prediction. For cohort C (Fig. 3b), nonventilator use, maximum serum glucose, and admission for medical reasons were the top indicators associated with in-hospital mortality. For cohort S and cohort G, there was a difference in the top variables between the models trained on datasets from different hospitals. Female sex and positive blood culture results were more important in cohort C (Fig. 3d), while a history of ulcers and community-acquired infection were more important in cohort G (Fig. 3f).
Critical variables with feature importance plots and SHAP values for predicting in-hospital mortality. Critical variables with feature importance plots and SHAP values for predicting in-hospital mortality. XGBoost extreme gradient boost, LightGBM light gradient boosting machine, SHAP Shapley additive explanation. (a) Feature importance including feature weight, mean gain and coverage of XGBoost and (b) SHAP value summary dot plot of the LightGBM-based prediction model in cohort C. (c) Feature importance including feature weight, mean gain and coverage of XGBoost and (d) SHAP value summary dot plot of the LightGBM-based prediction model in cohort S. (e) Feature importance including feature weight, mean gain and coverage of XGBoost and (f) SHAP value summary dot plot of the LightGBM-based prediction model in cohort G. The color of the SHAP dot represents the value of the feature, and the location of the dot on the X axis represents its SHAP value. Red dots indicate higher values or affirmative responses (for binary features), and blue dots indicate the opposite. A positive SHAP value indicates that the variables increase the likelihood of in-hospital mortality.
Discussion
Many hospitals and clinics already use EHR systems, and some of them have data extraction programs for analyzing big data that are accumulated in real time. ML analysis with these tools is much more convenient to use than traditional manual scoring systems, leverages the large amount of data already collected from EHRs to provide better predictive performance, and can even be automated to support clinical decision-making15,27,28. In this study, we demonstrated the ability of various ML algorithms to predict in-hospital mortality of ICU inpatients using EHR data and achieved improved performance compared with conventional predictive models.
Several previous studies had compared the predictive performance in prognosis of ICU patients, and various machine learning algorithms, including ANN, RF, and XGBoost were showed superior performance than conventional models15,16,18,29,30. In our study, all of the conventional models showed relatively low F1 scores (below 0.51), which is an informative index that evaluates binary classifiers on imbalanced datasets22,23. This might be because the conventional scoring systems failed to overcome the fundamental imbalance in clinical outcomes, where the in-hospital mortality rates of ICU inpatients included in cohort S and cohort G were 11.8% and 17.4%, respectively. In contrast, the F1 scores were largely improved in the ML models. In particular, the models trained with cohort S (the highest F1 score was 0.840), which consisted only of the data from hospital S, had a better fit than with cohort C (the highest F1 score was 0.765), which was constructed by combining data from the two hospitals. The conventional scoring system was modeled as a logistic regression method that could be affected by multicollinearity between variables, and the power of the model is expected to be reduced because it assumes that the relationship between all independent variables is linear31. ML-based models have inherent advantages for correcting nonlinear relationships and the multicollinearity of independent variables, and flexible updates are possible even when new clinical data are added. Thus, it has shown better performance in previous reports than models based on the conventional logistic regression method15,16,32.
Conventional models had moderate accuracy in predicting in-hospital mortality with our data, which is consistent with prior reports1,3,4,5,6,7. In particular, SAPS III and APACHE III had the highest AUROC among these models during the entire study period. Interestingly, the Pitt bacteremia score showed relatively higher performance with cohort G than with cohort S, which might be because the rate of cardiac arrest within 24 h of ICU admission, a variable included in the Pitt bacteremia score, was higher in hospital G than in hospital S. In our data, the optimal cutoff values with the highest AUROC increased over time in most models, possibly due to improved patient prognosis with the advances in health care systems and the extension of life expectancy. We applied updated optimal cutoffs for each study period to avoid the performance deterioration of conventional models seen in previous studies10,12,33,34,35,36.
We also demonstrated the top variables that had the largest impact on the predictive performance of the ML models in two university hospitals with differences in patient demographics, in-hospital mortality rate, and organizational practices. To predict in-hospital mortality in cohort C, ventilator application, glucose, type of admission, and blood culture results were selected as top predictors. However, the algorithm selected different features from the local data for cohort S and cohort G as the most predictive variables. ML models reflect more of the characteristics of each cohort and have a nontransferable and specific natures, as shown in the deteriorated external validation results of this study. Because each center has fundamentally different patient composition and characteristics, predictive models can be made more robust using ML approach trained on data from individual hospitals, but this inevitably leads to degrade performance on external data sets. Also in our study, algorithms such as LightGBM and XGBoost that performed best in the test set showed relatively more performance degradation in the external validation set. ML approach may have limitations in standardizing clinical predictive indicators or comparing the quality of care between centers18. In contrast, conventional scoring systems have relatively inferior predictive performance, but have strengths in standardization and center-to-center comparisons. Therefore, the use of ML models in conjunction with conventional scoring systems can provide more useful information for predicting the prognosis of critically ill patients or for comparing and evaluating ICUs.
Our study has several limitations. First, the models may not be generalized to other hospitals due to the single-country design of the study. In addition, residual confounding such as patient’s racial and socioeconomic status could act as algorithm bias. However, considering that the performance of the predictive model deteriorates over time, we attempted to demonstrate the importance of a system that can be updated continuously and can reflect the features of individual centers. Second, patients without demographic data or with more than 20% missing data were excluded from this study. Missing or inaccurate data may have affected the predictive power of the models. However, we attempted to minimize the impact of missing by using large data. It is expected that the improvement of EHR and data extraction tools can solve this problem in the near future.
Conclusion
We demonstrated the importance of an updatable predictive system that can reflect the characteristics of individual ICUs. Improvements in information technology and ML have opened new opportunities for analyzing clinical information. These techniques can be utilized to automatically and accurately collect patient data to support clinical decision-making using complex algorithms. Our study, employing models trained and validated on real-world data with two different cohorts, showed that ML-based models have accurate predictive power in the ICUs. In addition, conventional models have strengths in standardization and center-to-center comparisons. Using ML models in conjunction with conventional scoring systems can provide more useful information for predicting the prognosis of critically ill patients.
Data availability
The datasets generated and analysed during the current study are not publicly available but are available from the corresponding author on reasonable request.
References
De Mendonça, A., Bruining, H. & Thijs, L. The S0FA (Sepsis-related Organ Failure Assessment) score to describe organ. Intensive Care Med. 22, 707–710 (1996).
Tzouvelekis, L., Markogiannakis, A., Psichogiou, M., Tassios, P. & Daikos, G. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: An evolving crisis of global dimensions. Clin. Microbiol. Rev. 25, 682–707 (2012).
Knaus, W. A., Draper, E. A., Wagner, D. P. & Zimmerman, J. E. APACHE II: A severity of disease classification system. Crit. Care Med. 13, 818–829 (1985).
Le Gall, J.-R., Lemeshow, S. & Saulnier, F. A new simplified acute physiology score (SAPS II) based on a European/North American multicenter study. JAMA 270, 2957–2963 (1993).
Knaus, W. A. et al. The APACHE III prognostic system: Risk prediction of hospital mortality for critically III hospitalized adults. Chest 100, 1619–1636 (1991).
Zhu, B.-P. et al. Factors affecting the performance of the models in the Mortality Probability Model II system and strategies of customization: A simulation study. Crit. Care Med. 24, 57–63 (1996).
Higgins, T. L. et al. Updated mortality probability model (MPM-III). Chest 128, 348S (2005).
Moreno, R. P. et al. SAPS 3—From evaluation of the patient to evaluation of the intensive care unit. Part 2: Development of a prognostic model for hospital mortality at ICU admission. Intensive Care Med. 31, 1345–1355 (2005).
Henderson, H. et al. The pitt bacteremia score predicts mortality in nonbacteremic infections. Clin. Infect. Dis. 70, 1826–1833. https://doi.org/10.1093/cid/ciz528 (2020).
Patel, P. & Grant, B. Application of mortality prediction systems to individual intensive care units. Intensive Care Med. 25, 977–982 (1999).
Katsaragakis, S. et al. Comparison of acute physiology and chronic health evaluation II (APACHE II) and simplified acute physiology score II (SAPS II) scoring systems in a single Greek intensive care unit. Crit. Care Med. 28, 426–432 (2000).
Nassar, A. P. Jr. et al. Caution when using prognostic models: A prospective comparison of 3 recent prognostic models. J. Critical Care 27, 423 (2012).
Sakr, Y. et al. Comparison of the performance of SAPS II, SAPS 3, APACHE II, and their customized prognostic models in a surgical intensive care unit. Br. J. Anaesth. 101, 798–803 (2008).
Taylor, R. A. et al. Prediction of in-hospital mortality in emergency department patients with sepsis: A local big data–driven, machine learning approach. Acad. Emerg. Med. 23, 269–278 (2016).
Chiew, C. J., Liu, N., Wong, T. H., Sim, Y. E. & Abdullah, H. R. Utilizing machine learning methods for preoperative prediction of postsurgical mortality and intensive care unit admission. Ann. Surg. 272, 1133–1139. https://doi.org/10.1097/SLA.0000000000003297 (2020).
Kang, M. W. et al. Machine learning algorithm to predict mortality in patients undergoing continuous renal replacement therapy. Crit. Care 24, 42. https://doi.org/10.1186/s13054-020-2752-7 (2020).
Nanayakkara, S. et al. Characterising risk of in-hospital mortality following cardiac arrest using machine learning: A retrospective international registry study. PLoS Med. 15, e1002709. https://doi.org/10.1371/journal.pmed.1002709 (2018).
Roimi, M. et al. Early diagnosis of bloodstream infections in the intensive care unit using machine-learning algorithms. Intensive Care Med. 46, 454–462. https://doi.org/10.1007/s00134-019-05876-8 (2020).
Horan, T. C., Andrus, M. & Dudeck, M. A. CDC/NHSN surveillance definition of health care–associated infection and criteria for specific types of infections in the acute care setting. Am. J. Infect. Control 36, 309–332 (2008).
Calandra, T. & Cohen, J. The international sepsis forum consensus conference on definitions of infection in the intensive care unit. Crit. Care Med. 33, 1538–1548 (2005).
LeDell, E., Petersen, M. & van der Laan, M. Computationally efficient confidence intervals for cross-validated area under the ROC curve estimates. Electron. J. Stat. 9, 1583 (2015).
Saito, T. & Rehmsmeier, M. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS ONE 10, e0118432 (2015).
Chawla, N. V. Data mining for imbalanced datasets: An overview. Data Min. Knowl. Discov. Handbook 7, 875–886 (2009).
Lundberg, S. M. et al. Explainable machine-learning predictions for the prevention of hypoxaemia during surgery. Nat. Biomed. Eng. 2, 749–760 (2018).
Youden, W. J. Index for rating diagnostic tests. Cancer 3, 32–35 (1950).
Hosmer, D. W. Jr., Lemeshow, S. & Sturdivant, R. X. Applied logistic regression (Wiley, 2013).
Subudhi, S. et al. Comparing machine learning algorithms for predicting ICU admission and mortality in COVID-19. NPJ. Digit. Med. 4, 87. https://doi.org/10.1038/s41746-021-00456-x (2021).
Rajkomar, A., Dean, J. & Kohane, I. Machine learning in medicine. N. Engl. J. Med. 380, 1347–1358 (2019).
Kim, S., Kim, W. & Park, R. W. A comparison of intensive care unit mortality prediction models through the use of data mining techniques. Healthcare Inf. Res. 17, 232–243 (2011).
Hsieh, M. H. et al. Comparison of machine learning models for the prediction of mortality of patients with unplanned extubation in intensive care units. Sci. Rep. 8, 1–7 (2018).
Baxt, W. G. Complexity, chaos and human physiology: The justification for non-linear neural computational analysis. Cancer Lett. 77, 85–93 (1994).
Gould, M. K., Huang, B. Z., Tammemagi, M. C., Kinar, Y. & Shiff, R. Machine learning for early lung cancer identification using routine clinical and laboratory data. Am. J. Respir. Crit. Care Med. 204, 445–453. https://doi.org/10.1164/rccm.202007-2791OC (2021).
de Silva, C. V. T. et al. Performance of the third-generation models of severity scoring systems (APACHE IV, SAPS 3 and MPM-III) in acute kidney injury critically ill patients. Nephrol. Dial. Transpl. 26, 3894–3901 (2011).
Soares, M. et al. Validation of four prognostic scores in patients with cancer admitted to Brazilian intensive care units: Results from a prospective multicenter study. Intensive Care Med. 36, 1188–1195 (2010).
Wu, V. C. et al. Patients supported by extracorporeal membrane oxygenation and acute dialysis: acute physiology and chronic health evaluation score in predicting hospital mortality. Artif. Organs 34, 828–835 (2010).
Maccariello, E. et al. SAPS 3 scores at the start of renal replacement therapy predict mortality in critically ill patients with acute kidney injury. Kidney Int. 77, 51–56 (2010).
Author information
Authors and Affiliations
Contributions
M.H.C., Y.J.C., J.H.C. and S.H.J. contributed to the design of the study. M.H.C. and D.K. collected the data. M.H.C., E.J.C. and Y.J.J. analyzed and interpreted the data. M.H.C. and S.H.J. drafted the manuscript. D.K., Y.J.C., J.H.C. and S.H.J. reviewed the manuscript. All authors read, commented, and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Choi, M.H., Kim, D., Choi, E.J. et al. Mortality prediction of patients in intensive care units using machine learning algorithms based on electronic health records. Sci Rep 12, 7180 (2022). https://doi.org/10.1038/s41598-022-11226-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-11226-4
- Springer Nature Limited