Abstract
Among bacterial species implicated in hospital-acquired infections are the emerging Pan-Drug Resistant (PDR) and Extensively Drug-Resistant (XDR) Acinetobacter (A.) baumannii strains as they are difficult to eradicate. From 1600 clinical specimens, only 100 A. baumannii isolates could be recovered. A high prevalence of ≥ 78% resistant isolates was recorded for the recovered isolates against a total of 19 tested antimicrobial agents. These isolates could be divided into 12 profiles according to the number of antimicrobial agents to which they were resistant. The isolates were assorted as XDR (68; 68%), Multi-Drug Resistant (MDR: 30; 30%), and PDR (2; 2%). Genotypically, the isolates showed three major clusters with similarities ranging from 10.5 to 97.8% as revealed by ERIC-PCR technique. As a resistance mechanism to fluoroquinolones (FQs), target site mutation analyses in gyrA and parC genes amplified from twelve selected A. baumannii isolates and subjected to sequencing showed 12 profiles. The selected isolates included two CIP-susceptible ones, these showed the wild-type profile of being have no mutations. For the ten selected CIP-resistant isolates, 9 of them (9/10; 90%) had 1 gyrA/1 parC mutations (Ser 81 → Leu mutation for gyrA gene and Ser 84 → Leu mutation for parC gene). The remaining CIP-resistant isolate (1/10; 10%) had 0 gyrA/1 parC mutation (Ser 84 → Leu mutation for parC gene). Detection of plasmid-associated resistance genes revealed that the 86 ciprofloxacin-resistant isolates carry qnrA (66.27%; 57/86), qnrS (70.93%; 61/86), aac (6')-Ib-cr (52.32%; 45/86), oqxA (73.25%; 63/86) and oqxB (39.53%; 34/86), while qepA and qnrB were undetected in these isolates. Different isolates were selected from profiles 1, 2, and 3 and qnrS, acc(6,)-ib-cr, oqxA, and oqxB genes harbored by these isolates were amplified and sequenced. The BLAST results revealed that the oqxA and oqxB sequences were not identified previously in A. baumannii but they were identified in Klebsiella aerogenes strain NCTC9793 and Klebsiella pneumoniae, respectively. On the other hand, the sequence of qnrS, and acc(6,)-ib-cr showed homology to those of A. baumannii. MDR, XDR, and PDR A. baumannii isolates are becoming prevalent in certain hospitals. Chromosomal mutations in the sequences of GyrA and ParC encoding genes and acquisition of PAFQR encoding genes (up to five genes per isolate) are demonstrated to be resistance mechanisms exhibited by fluoroquinolones resistant A. baumannii isolates. It is advisable to monitor the antimicrobial resistance profiles of pathogens causing nosocomial infections and properly apply and update antibiotic stewardship in hospitals and outpatients to control infectious diseases and prevent development of the microbial resistance to antimicrobial agents.
Similar content being viewed by others
Introduction
A. baumannii is a strictly aerobic bacteria, non-lactose-fermenter, Gram-negative, and an opportunistic pathogen, which causes hospital-acquired infections1. The infection caused by this organism is difficult to treat2 due to its resistance to different classes of antimicrobial agents that is attributed to the intrinsic resistance and the organism ability to acquire resistance determinants as a result of the its genome plasticity. This endless capacity of genetic variability is the reason behind the global emerging problem caused by this organism3.
Different acquired resistance mechanisms as a result of overuse by physicians or misuse of antibiotics by patients have been reported for this pathogen and therefore, causing it to be able to express PDR or extensively drug-resistant (XDR) phenotypes particularly among critically ill patients4. Fluoroquinolones (FQs) in the last four decades had shown good activity against A. baumannii isolates. However, resistance to these drugs has rapidly emerged4. FQs are a widely prescribed medication in Egypt, and quinolone resistance has jumped up sharply5. A baumannii is now non-susceptible to the greatest repertoire of antimicrobial agents, involving FQs, and Pan-Drug resistant (PDR) is often responsible for the miscarriage of antibiotic treatment6. Resistance to FQs is primarily caused by the spontaneous mutations of genes in the area of the quinolone resistance-determining region (QRDR), which includes DNA gyrase, and topoisomerase IV. Changes that affect the drug to exert its goal due to alterations in the drug target genes, DNA gyrase (gyrA) or Topoisomerase IV C (parC) subunits, have been related to high levels of resistance to FQs7.
However, Martinez has detected the prevalence of plasmid-associated fluoroquinolones resistance (PAFQR) genes more than two decades ago8. Despite their insufficiency to confer FQ resistance, PAFQR executes an important role in the procuration of resistance to FQ by facilitating the selection of additional chromosomal resistance mechanisms, leading to a high level of FQ resistance and enabling bacteria to become fully resistant9. Most importantly, PAFQR can spread horizontally among A. baumannii10. Three kinds of PAFQR determinants have been described; qnr which protects fluoroquinolones targets from inhibition8. Inactivation of fluoroquinolones by acetylation with the common aminoglycoside acetyltransferase aac (6)-Ib-cr11, and efflux pumps QepAB and OqxAB12 are among the resistance mechanisms exhibited by this organism. Few studies were published about the prevalence of PAFQR determinants among A. baumannii isolates13. Touati, 2008 reported the detection of qnrA in A. baumannii in Algerian hospitals for the first time14. Jiang, 2014 reported the detection of two qnrB-positive isolates of PAFQR in clinical isolates of A. baumannii from Henan hospital, China15. A publication from China has also reported the prevalence of qnrB6 and qnrS213. Many studies have recently reported the emergence of resistance of A. baumannii in Egypt, limited studies are available on the mechanisms responsible for the resistance of A. baumannii in Upper Egypt to fluoroquinolones. Therefore, this study aimed to determine the antibiotic-resistant pattern of A. baumannii recovered from nosocomial infection cases and FQs-resistant mechanisms as an attempt to control this life-threatening pathogen.
Materials and methods
Specimens collection
A total of 1600 specimens were collected from blood, respiratory tract, urinary tract, catheters, wounds, and skin between January 2014 and March 2019 from two university hospitals; Al Azhar university hospital and Assuit university hospital, Assuit governorate, upper Egypt. Different clinical isolates were recovered. The study was approved by the ethics committee (Ethics Committee ENREC-ASU-63) of faculty of pharmacy-Ain Shams university. Both informed and written consents were obtained from the patients after explaining the study purpose. Also, all methods were performed following the relevant guidelines and regulations. The isolates were identified by standard microbiological methods16 and as described below.
Phenotypic and genotypic identification of A. baumannii isolates
All specimens were cultured on nutrient agar (Oxoid Limited, England) as a general medium to recover all bacterial pathogens. The recovered bacterial isolates were characterized using two selective media, MacConky agar (Oxoid Limited, England) to identify lactose fermenting from non-lactose fermenting species and Herellea agar (Himedia, India) for selective scoring of Acinetobacter spp. Incubation was done at 37 °C for 24 h. All collected clinical isolates were propagated and maintained by standard microbiologic techniques16. For phenotypic identification, single separate colonies were handled for qualitative conventional diagnostic tests for A. baumannii; including Gram staining. The typical isolates showed Gram-negative reaction, catalase, and citrate utilization positive, while oxidase and indole tests were negative17. For genotypic identification, genomic DNA was extracted using the GeneJET Genomic DNA Purification Kit (Thermo, USA, catalog No. K0721) and used for amplification of the intrinsic blaOXA-51-like gene. This gene is unique for A. baumannii as previously described18. A. baumannii ATCC 19,606 was used as a positive control.
Antimicrobial susceptibility testing
Disk diffusion was carried out on 19 antimicrobial agents using the Kirby-Bauer disk diffusion method as recommended by the Clinical and Laboratory Standards Institute (CLSI)19. The antibiotic discs used for susceptibility testing were imipenem (IMP 10 μg), meropenem (MEM 10 μg), piperacillin (PRL 100 μg), piperacillin/tazobactam (TZP 100/10 μg), ampicillin/sulbactam (SAM 10/10 μg), ceftazidime (CAZ 30 μg), cefotaxime (CTX 30 μg), ceftriaxone (CRO 30 μg), cefepime (FEP 30 μg), amikacin (AMK 10 μg), tobramycin (TOP 10 μg), gentamicin (CN 10 μg), ciprofloxacin (CIP 5 μg), levofloxacin (LEV 5 μg), gatifloxacin (GAT 5 μg), trimethoprim/sulfamethoxazole (SXT 12.5/23.75 μg), tigecycline (TGC15 μg), doxycycline (DO 30 mcg) and colistin (CT 10 units). All antimicrobial discs were purchased from Oxoid (UK) except gatifloxacin discs were purchased from Himedia (India). MDR, XDR, and PDR phenotypes were identified as previously determined20.
Molecular typing of CIP-resistant A. baumannii isolates
Investigation of clonal relationship and diversity of the recovered A. baumannii isolates was determined by molecular typing of the CIP-resistant isolates using ERIC-PCR21. Genomic DNA was extracted using the Genomic DNA Purification Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. ERIC-PCR was carried out using the ERIC-1 (5’-ATGTAAGCTCCTGGGGATTCAC-3’) and ERIC-2 (5’-AAGTAAGTGACTGGGGTGAGCG-3’) primers as previously described21. The PCR products were analyzed using agarose gel electrophoresis at 1.5% (w/v) agarose containing 0.5 mg/ml ethidium bromide that was subsequently visualized by UV transilluminator. ERIC-PCR dendrogram was constructed by the use of UPGMA clustering method, Bionumeric program version 7.6 (Applied Maths). The percentage of similarity among the 86 CIP-resistant A. baumannii isolates was calculated by the use of Jaccard's Coefficient22.
Molecular characterization of FQs resistance mechanisms in A. baumannii isolates
Detection of target-site mutation in genomic DNA
Genomic DNA was extracted using the GeneJET Genomic DNA Purification Kit (Thermo, USA, catalog No. K0721).
-
A.
PCR screening of FQ-resistance target site mutation
PCR condition denaturation at 95 °C for 2 min, then 35 cycles of amplification as follows: denaturation at 95 C for 30 s, annealing for 30 s at primer set-specific temperatures, and extension at 72 °C for 1 min), a final extension at 72 °C for 5 min. The oligonucleotides used to amplify and for sequencing analysis are shown in Table 123.
-
B.
Purification of PCR products
Purification of PCR products before sequencing was performed using the PCR purification kit (Thermo, USA, Catalog number: K0701). Briefly, 1 volume of DNA binding buffer was mixed with each volume of PCR product. The sample mixture was then transferred into thermo-spin and centrifuged. The column was washed with a DNA wash buffer. Finally, the column was eluted with 50 µl DNA elution buffer. The purified DNA was stored at -200C for safe storage of amplicons till sequencing using Sanger sequencing technique (Applied Biosystems genetic analyzers (ThermoFisher, Uk)).
-
C.
Target site sequence analysis
A 613 bp and A 919 bp fragments of the corresponding gyrA and parC genes were amplified from 12 selected isolates (one isolate from one out of 12 profiles, isolates of the profiles 1 to 12 showed resistance to 19, 18, 17, 16, 15, 14, 13, 12, 11, 9, 8, 6 out of 19 tested antimicrobial agents, respectively. The chosen 12 profiles included PDR XDR and MDR isolates). Nucleotide sequences were visualized by SnapeGene Viewer (version 5.1.4.1 software 2020). ORFs were identified using the ORF finder tool. The sequences of gyrA and parC genes from the selected 12 isolates were compared to their corresponding ones of the gyrA and parC genes of A. baumannii ATCC 19,606 as previously described24. Pairwise codon-based nucleotide alignments (CDS-alignments) of the gyrA and parC genes from the selected isolates against their corresponding sequences of the standard strain were carried out.
Plasmid-associated fluoroquinolone resistance (PAFQR) genes
-
A.
Plasmid DNA extraction
Plasmid DNA was extracted and purified using the GeneJET Plasmid Miniprep Kit (Thermo, USA, catalog No. K0502). Briefly, a single colony from each test isolates was picked up from a freshly streaked selective plate to inoculate 1–5 mL of Mueller Hinton broth mixed with ciprofloxacin (0.125 mg/ml) in a test tube. The tubes were incubated at 37 °C for 12–16 h under shaking at 200–250 rpm in a shaker incubator. The volume of the container either a test tube or a flask was at least 4 times the culture volume. Cell pellets of the bacterial culture were collected by centrifugation at 6800 × g (8000 rpm) for 2 min using a micro-centrifuge. The supernatant was decanted and bacterial pellets were resuspended and subjected to alkaline lysis to liberate the plasmid DNA. The resulting lysate was neutralized to create appropriate conditions for the binding plasmid DNA on the silica membrane in the spin column. By centrifugation, cell debris was pelleted, and the supernatant containing the plasmid DNA was loaded onto the spin column membrane. The adsorbed DNA was washed to remove contaminants and then eluted with 50 µL of the elution buffer (10 mM Tris–HCl, pH 8.5). The extracted plasmid DNA was stored at − 20 °C for subsequent use.
-
B.
PCR screening of PAFQR determinant genes
Plasmid extracts of all isolates were screened for the following genes: qnrA, qnrB, qnrS, aac(6′)-Ib-cr, qepA, oqxA , and oqxB using gene-specific primers listed in Table 2. PCR conditions and primer sequences were as previously described25.
-
C.
Sequencing of resulting amplicons of target genes
PCR amplicons of target genes: qnrA, qnrB, qnrS, aac(6′)-Ib-cr, qepA, oqxA , and oqxB resulting from plasmid extracts of test isolates from selected profiles numbers 1, 2, and 3 only were sequenced using Sanger sequencing technique (Applied Biosystems genetic analyzers (ThermoFisher, UK)). Such kind of isolates might be more life-threatening ones.
Statistical analysis
All data were analyzed using the GraphPad PRISM (Version.8.4.0.671). Descriptive statistics were used. The p-value < 0.05 was considered a statistically significant used Chi-Square test.
Ethical approval
The study was approved by the Ethics Committee (Ethics committee ENREC-ASU-63) at Faculty of Pharmacy-Ain Shams University where both informed and written consent were obtained from the patient after explaining the study purpose.
Results
Phenotypic and genotypic identification of A. baumannii isolates
The isolates were identified according to standard microbiological methods16. The isolates showing characters suspected to be Acinetobacter species were subjected to some biochemical and growth conditions tests. The suspected isolates of Acinetobacter spp gave the following test results: negative reaction with oxidase, and indole, positive reaction with catalase test, growth at 44 °C, and positive for citrate utilization test. Examination of cell morphology of Gram-stained films revealed that: all Acinetobacter isolates were short, Gram-negative diplo-coccobacillus (sometimes de-staining of primary stain was difficult due to tendency of some isolates to retain crystal violet) with pairing or clustering arrangements. From a total of 623 non-lactose fermenters isolates, only 151 had these mentioned characters.
All Acinetobacter species candidates (151 isolates) were further subjected to conventional PCR to confirm their identity. The characteristic band at 353 bp for the blaOXA-51-like gene of A. baumannii was used for this identification and confirmation. From the total 151 isolates subjected to PCR, only 100 isolates were positive for blaOXA-51 and confirmed to be A. baumannii as shown in Supplementary Fig. S1.
The specimens that showed the highest percentage 61% (61/100) of A. baumannii contamination were obtained from the respiratory tract (ETT 29%, nasal 17%, sputum 13%, throat 2%), followed by urinary tract infection 17% (urine 9%; urinary tract catheter 8%) and blood 12%, while the lowest (2%; 2/100) was from skin and CVC.
Antimicrobial susceptibility testing
As shown in Table 3 all A. baumannii isolates exhibited high resistance to most of the tested antimicrobial agents. However, the higher resistance was recorded to piperacillin (99%) and cephalosporins (98%). On the other hand, the most effective antimicrobial agents were recorded to be colistin (only 5% of isolates showed resistance) followed by doxycycline (only 57% of isolates showed resistance).
Resistance profile analysis of A. baumannii susceptibility results (100 isolates) revealed that two isolates (2%) were detected as pan drug-resistant (PDR), 68% isolates were extensive drug-resistant (XDR), and 30% isolates were multidrug-resistant (MDR).
Analyses of the resulting A. baumannii susceptibility to the 19 tested antimicrobial agents showed the diversity of isolates' resistance to the tested agents. They were divided into 12 profiles according to the number of antimicrobial agents to which they were resistant. The resistance scope ranged from 6 to 19 antimicrobial agents. The first profile is PDR (two isolates), which represents isolates resistant to 19 tested antimicrobial agents. The second profile is XDR (32 isolates), which represents isolates resistant to all tested antimicrobial agents except colistin, and the third profile (19 isolates) represents isolates resistant to 17 antimicrobial agents. On the other hand, profiles numbers 4 to 12 represent A. baumannii isolates susceptible to 3 to 13 of the 19 tested antimicrobial agents (Fig. 1).
Molecular typing of CIP-resistant A. baumannii isolates
Determination of clonal relationship and diversity of hospital-acquired infection caused by A. baumannii isolates were carried out by the genotyping method using ERIC-PCR technique. This was conducted for all A. baumannii isolates resistant to CIP (n = 86), and the results were subjected to phylogenetic analysis (Fig. 2). As a result, the isolates could be divided into three major clusters (I, II, and III) with similarities ranging from 10.5 to 97.8%. The high diversity indicates multiple contamination sources with A. baumannii. Cluster I represents 63.95% (55/86). Cluster II represents 2.32%, and included only two isolates namely; AS-44 and AS-46, while cluster III constitutes 37% (29/86) of the total isolated A. baumannii (Fig. 2). All A. baumannii in clusters II and III were recovered from Assiut University while most isolates included in cluster I was recovered from Al-Azhar University.
Molecular characterization of A. baumannii resistance mechanisms to FQs
Detection of target site mutation
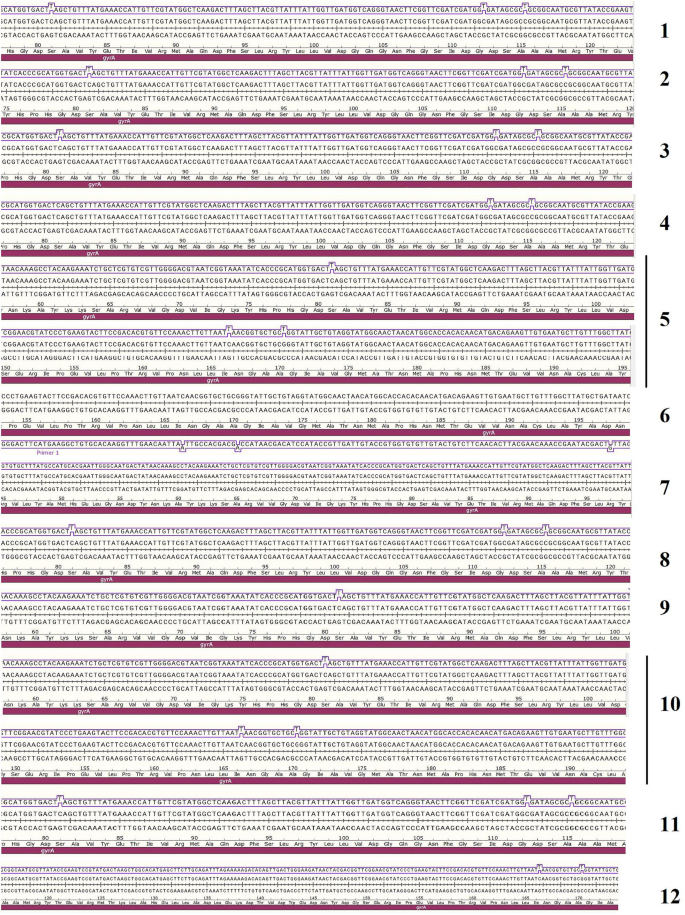
Out of 12 phenotypic profiles (Fig. 1), one isolate from each profile was selected, and its purified genome was subjected to PCR amplification of gyrA and parC genes followed by sequencing of the resulting amplicons. Out of the 12 isolates, 10 isolates were fluoroquinolone-resistant, and two isolates were fluoroquinolone susceptible (AS-01 and AS-05). gyrA and parC gene sequences of the selected A. baumannii isolates were analyzed and they showed similarities ranging from 95 to 100% to their corresponding sequences deposited in GenBank nucleotide database under accession numbers shown in Supplementary Table 1. Target site mutation analyses in gyrA and parC gene sequences of the selected twelve A. baumannii isolates represent 12 profiles. Two isolates (susceptible ones to CIP) had a wild-type profile. For the ten isolates (CIP-resistant ones), 9 of them (9/10; 90%) had 1 gyrA and 1 parC mutations Ser 81 → Leu mutation for gyrA gene and Ser 84 → Leu mutation for parC gene. The remaining CIP-resistant isolate (1/10; 10%) had (0 gyrA /1 parC) mutation (Ser 84 → Leu mutation) for parC gene. All tested isolates had a silent mutation in one or more positions of either gyrA or parC or both gyrA and parC (Supplementary Table 2, and Figs. 3 and 4).
Plasmid-associated fluoroquinolone resistance (PAFQR) genes
-
A.
DNA plasmid extraction
The variable number of bands per isolate may be plasmids with different molecular weights that were detected (no endonuclease digestion was used) in 99% of A. baumannii. Besides, no plasmid could be detected in 1% of the isolates.
-
B.
PCR screening A. baumannii isolates for PAFQR genes
All isolates were screened for PAFQR genes (qnrA, qnrB, qnrS, acc(6)-ib, qepA, oqxA, and oqxB) using conventional PCR. The expected sizes of PCR products mentioned in Table 2 were obtained. Ciprofloxacin-resistant isolates (86%; 86/100) carried qnrA (66.27%; 57/86), qnrS (70.93%; 61/86), aac (6')-Ib-cr (52.32%; 45/86), oqxA (73.25%; 63/86) and oqxB (39.53%; 34/86) resistance genes, while the resistance genes qepA and qnrB were undetected in these isolates. Although 14 isolates were susceptible to ciprofloxacin, some resistant genes were detected in these isolates, these included qnrA (7/14), qnrS (7/14), aac (6')-Ib-cr (3/14), oqxA (12/14), and oqxB (11/14).
-
C.
Distribution of PAFQR genes among A. baumannii isolates
-
(i)
Among CIP-resistant isolates
Analysis of the 86 CIP-resistant isolates for their acquisition of PAFQR genes gave 26 profiles (Table 4). The prevalence of PAFQR genes was highly observed among 84 isolates. The plasmid extracts of isolates might contain up to five genes per isolate. On the other hand, the plasmid extract of one isolate (AS-29) did not show any resistance gene and another isolate (AZ-18) which did not carry any plasmid. The latter isolate was recovered from the sputum of an admitted patient who stayed for only three days in Al-Azhar university hospital.
-
(ii)
Among CIP-sensitive isolates (n = 14)
For the 14 CIP-sensitive isolates, analyses of occurrence and distribution of PAFQR genes in their corresponding plasmid extracts revealed that these plasmids harbor up to five genes, which gave 8 profiles of genes association (Fig. 5). On the other hand, one isolate did not harbor any PAFQR resistance gene.
-
(iii)
PAFQR gene sequences analyses
PCR products of different genes from an isolate representing each profile 1, 2, and 3 were selected for sequencing (contain more life-threatening isolates), these included qnrA, qnrS, acc(6,)-ib-cr, oqxA, and oqxB. The amplicon sizes were 347, 255, 480, 489, and 480 bp, respectively. The BLAST of the NCBI (www.ncbi.nlm.nhi.gov) was used to search databases for detecting the similarity in nucleotides and amino acid sequences of these studied genes to those deposited in the databases. The BLAST results revealed that the oqxA and oqxB sequences were not identified previously in A. baumannii but they were identified in Klebsiella aerogenes strain NCTC9793 and Klebsiella pneumoniae, with an identity of 99.78% and 99.77%, respectively. On the other hand, the sequences of qnrA, qnrS, acc(6,)-ib-cr, and oqxA, showed homology to those of A. baumannii deposited in GeneBank database with identity ranged from 97.98 to 98.28% (Table 5).
Discussion
A. baumannii is a Gram-negative bacterium, can withstand a wide range of environmental conditions and also can survive on surfaces. These characters enable it to be implicated in many nosocomial infections and outbreaks. It is a strict aerobic organism2. The infection caused by A. baumannii is difficult to treat2. It has been recognized by the Infectious Disease Society of America as one of the six highly drug-resistant hospital pathogens26.
Antimicrobial agents use strategies are known to significantly reduce the frequency of bacterial infections in patients27. Among which fluoroquinolones (FQs) have the widest use and are currently recommended by different physicians as they have multiple applications and different advanced generations4. Nevertheless the emergence and spread of bacterial resistance to FQs among Gram-negative bacteria generally and A. baumannii specifically is becoming increasingly serious with their extensive use. FQs in the last forty years had shown good activity against A. baumannii isolates, However, resistance to these drugs has rapidly emerged15. FQs are widely prescribed medication in Egypt, and resistance to FQs has skipped pointedly5. The developing resistance of A. baumannii to antimicrobial agents has been described and this was attributed to the abundance of these antibiotics in multiple pharmaceutical markets28, besides their misuse29. A. baumannii infection is difficult to remedy, as of its everlasting fullness to acquire antimicrobial resistance due to the suppleness of its genome30. Many acquired resistance mechanisms have been reported for this pathogen and therefore, render it able to express MDR, XDR, or PDR phenotypes that were associated with significant morbidities and mortalities4. Therefore, this study aimed to determine the mechanisms behind A. baumannii resistance to FQs. For achieving this aim, the following objectives were studied: (i) the antibiotic-resistant phenotypes of A. baumannii recovered from nosocomial infection cases in Al-Azhar university hospital and Assiut university hospitals, both in Assiut governorate Upper Egypt; (ii) studying the molecular mechanisms responsible for the resistance of A. baumannii isolates to FQs which included target site mutation and ESBLs as well as PAFQR genes acquisition.
In this study, a total of 1600 specimens were collected from different clinical hospitalized patients of two major university hospitals in Upper Egypt during the period between January 2014 and March 2019. Out of 151 Acinetobacter candidate isolates subjected to PCR, only 100 isolates were positive for blaOXA-51 and confirmed to be A. baumannii. The identification of A. baumannii phenotypically is difficult due to significant phenotypic overlapping with other species, which are genotypically closely related to each other31.
The study results revealed that the highest number of A. baumannii isolates was recovered from respiratory and urinary tracts as well as blood specimens indicating the involvement of this pathogen in infection of these sites. The 100 recovered A. baumannii isolates comprised 61 isolates from respiratory tract infection {Enotreacheal tubes (29), nasal (17), sputum (13), and throat (2)}, blood infection (12), and urinary tract infections (9). It has been reported32 that the respiratory tract, blood, and urinary tract constitute the most predominant sources of A. baumannii pathogen. A. baumannii was also recovered from wounds or soft-tissue infections (6), skin (2), catheter-associated infections {central venous catheter (2), and urinary tract catheter (8)} in agreement with the results reported by33. Worldwide, it was reported that A. baumannii infection differs according to both the anatomical site and the clinical conditions of the patients34.
FQs in the last four decades had shown good activity against A. baumannii isolates, however, resistance to these drugs has quickly been rised15. FQs are a widely prescribed medication in Egypt, and FQs resistance has jumped up sharply4, 5, 35. Our findings emphasize that most A. baumannii isolates (71%) were recovered from patients treated for long period with FQs, while the rest (29%) of A. baumannii isolates were recovered from patients treated with other antimicrobial agents such as cephalosporins, imipenem, and the penicillin derivative (amoxicillin) or combination (amoxicillin-clavulanic acid). Un-rational use, low dose, and misuse of antimicrobial agents (empirical antibiotcs use by non hospitalized patients) result in the development of microbial resistance and so increase the risk of nosocomial Acinetobacter infections in hospitals4.
The increase in A. baumannii resistance constitutes a global issue36. Every year, the life of millions of hospitalized patients are seriously affected by incurable strains of A. baumannii37. As an infection control measure, continuous studies on the resistance profile of this organism are highly required and are a must for at least decreasing its devastating effect on the quality of medical treatment4. In the current study, the prevalence of resistance among the 100 recovered A. baumannii isolates against the tested antimicrobial agents was high. A resistance prevalence of ≥ 78% was recorded for the tested isolates against the 19 antimicrobial agents used. On the other hand, colistin proved to be the most effective anti-microbial agent against these isolates (95% of isolates were sensitive) followed by doxycycline (43% of isolates were sensitive). Our finding agrees with previous studies which stated that, A. baumannii pathogen is an opportunistic organism, often susceptible to colistin and having a low susceptibility to other antimicrobial agents. This organism is involved in radical morbidity and mortality38.
The emerged pan drug-resistant (PDR), extensively drug-resistant (XDR)-A. baumannii strains could be a leading cause of hospital-acquired infections by this opportunistic pathogen4. PDR and XDR are being recorded increasingly among A. baumannii isolates recovered from clinical 4, 39 or environmental (such as soil) specimens40. In our finding, two isolates (2%) were detected as PDR, 68% isolates were XDR and 30% isolates were MDR. PDR and most of XDR- A. baumannii were isolated from Assiut university hospitals. The prevalence of MDR, XDR, and PDR A. baumannii could be attributed to the misuse of antimicrobial agents41, 42, or due to the differences in rates of infections by the respective pathogens (mostly related to the degree of strict hygiene protocols applied in different hospitals)43, in addition to the plasticity and endless capacity of changes demonstrated in A. baumannii genomes44.
Genotyping method using ERIC-PCR technique was applied for determining the clonal relationship and diversity of isolated A. baumannii used in the present study. This technique can be used during nosocomial outbreaks to investigate if the involved isolates are genetically related or originated from the same strains45. The use of strain typing in infectious disease control decisions in hospitals is based on several assumptions, (i) whether the isolates associated with an outbreak are the progeny of a single clone, or (ii) have identical genotype, or (ii) epidemiologically unrelated so have different genotypes46. In our study, the phylogenetic dendrogram of ERIC-PCR showed that the isolates can be divided into three major clusters. This diversity might be due to multiple contamination sources by this organism, a finding that is different from that reported by some authors47 who showed clonal expansion and microbial colonization by the Acinetobacter baumannii isolates used in their study. The obtained results necessitate the continuous monitoring of emerged genotypes among bacterial species implicated in hospital acquired infection and the development of new infection control strategies to comate the spread of such pathogens.
In the current study, investigation of the mechanisms responsible for the resistance of A. baumannii isolates to FQs involved detection of target-site mutation occurrence and acquisition of PAFQR genes. The analyses of target site mutations in gyrA and parC gene sequences of twelve selected A. baumannii isolates showed 12 profiles. Two isolates were susceptible to CIP and had a wild-type profile of being have no mutations. Ten isolates were CIP-resistant, 9 of them (9/10; 90%) had (1 gyrA /1 parC) mutation: Ser 81 → Leu mutation for gyrA gene and Ser 84 → Leu mutation for parC gene. The remaining CIP-resistant isolate (1/10; 10%) had (0 gyrA /1 parC) mutation: Ser 84 → Leu mutation for parC gene as reported previously in North Egypt48. Previous studies stated that double mutations (1gyrA/1parC) could be sufficient to confer CIP resistance in A. baumannii isolates23, 48, 49 and this in agreement with our results. . It was reported that these two mutations are enough to predict resistance to CIP and LEV48. However, gyrA and parC mutations did not occur in all resistant mutant strains and the resistance may be attributed to changes in outer membrane protein expression and drug efflux pumps50. All tested isolates had a silent mutation in one or more positions of either gyrA or parC or even both gyrA and parC that do not lead to a change in amino acid composition. Twenty-three isogenic mutations in gyrA genes were as follows: (6 Gly112, 7 Ala115, 4 Ile166, 4 Ala170, 2 Asp197) and eleven isogenic mutations in parC gene as follows: (2 Ala 52, 6 Leu 35, 1 Ala 127, 1 Gly 143, 1 Ala163). The isogenic mutations were identified previously in Gram-negative51,52,53, Gram-positive54, 55, Mycobacterium56, Mycoplasma57, and A. baumannii58. Our findings showed that four amino acid affected by isogenic mutations were detected in gyrA, these included Ala, Asp, Gly, and Ile while the three amino acid-affected by isogenic mutations in parC included Ala, Gly, and Ile and occurred in different codon positions. A storm of silent mutations was identified previously in A. baumannii50. Isogenic or silent mutations may be due to that the patient's administrated insufficient doses of FQs in different periods which may induce bacterial resistance to the drug58.
The dissemination of mobile elements harboring resistance genes between Acinetobacter spp. is not fully understood. Many earlier investigations have given special emphasis to plasmid-mediated horizontal transfer of antibiotic resistance genes59,60,61. In the present study, 99%; 99/100 of A. baumannii have harbored plasmids. However, one isolate that represents 1% has not harbored any plasmids. Despite plasmids themselves may be insufficient to confer FQs resistance, PAFQR genes execute an important role in the procuration of resistance to FQs by facilitating the selection of additional chromosomal resistance mechanisms, leading to a higher level of quinolone resistance and enabling bacteria to become fully resistant62. Most importantly, PAFQR genes can spread horizontally among A. baumannii10. Resistance to quinolones can be mediated by plasmids63. Three kinds of PAFQR determinants have been described: qnr genes (qnrA, qnrB, and qnrS) that encode pentapeptide protein repeats, which protect the quinolone targets from inhibition8, 63. Inactivation of fluoroquinolones occurs by acetylation with the common aminoglycoside acetyltransferase aac (6′)-Ib-cr11 and can be pumped out by efflux pumps QepAB and OqxAB12. The first PAFQR gene type is qnr genes, the 86, ciprofloxacin-resistant isolates carried qnrA (66.27%; 57/86), qnrS (70.93%; 61/86), while qnrB was undetected in these isolates. Such a high prevalence reported here and in other studies performed worldwide among FQ-resistant A. baumannii isolates reflects their fetal role in the acquisition of FQs resistance genes. In disparity, the lower prevalence of qnr was reported in CIP-resistance A. baumannii isolates from Brazil (37.5%) and north Egypt 48.3%64. On the other hand, qnr genes were more frequently detected among the isolates of K. pneumoniae (70.4%) and E. coli (67.5%)61, 62, 65, 66. The second PAFQR gene type is the aac(6')-Ib-cr gene, a new variant of common aminoglycoside acetyltransferase that acetylates piperazinyl substituent of some fluoroquinolones, including ciprofloxacin11. The bifunctional aminoglycoside and fluoroquinolone active variant aac (6')-Ib-cr catalyzes the acetylation of both drug classes67. PCR screening of isolates tested in the present study showed the prevalence of aac (6')-Ib-cr (52.32%; 45/86) among CIP-resistance isolates. On the other hand, CIP-susceptible isolates harbored aac (6')-Ib-cr by only 21.42% (3/14), these isolates were already resistant to aminoglycosides. Our result agrees those obtained in Iran and Brazil68, 69 but disagrees with that of Hamed et al.25 in north Egypt. The third PAFQR gene types are those encoding the efflux pumps QepA, and oqxAB12. QepA is a proton-dependent transporter belonging to the major facilitator superfamily that causes hydrophilic quinolone resistance12, while OqxAB is a transmissible resistance-nodulation-division multidrug efflux pump that was found to reduce susceptibility to CIP and nalidixic acid70. PCR screening showed the prevalence of oqxA by 73.25% (63/86) and oqxB by 39.53% (34/86) while qepA was undetected in these isolates. Our results don’t agree with that reported by Hamed et al.25 since they were unable to detect oqxAB in A. baumannii and E. coli while they could detect both in Klebsiella spp25. qepA was not detected in A. baumannii , E. coli, and K. pneumoniae but detected in Enterobacter spp. in the study conducted by71. Summing up, high variation in the prevalence of PAFQR efflux genes among different microbial species significantly limits the treatment options of infected patients and provides a potential source for the horizontal spread of resistance71. The differences mentioned above can result from the geographical distance, surveillance strategies, and variability in following up antibiotic stewardship among organizations.
Our findings showed that, although some of A. baumannii isolates (14%; 14/100) were CIP-sensitive, they harbored PAFQR genes: qnrA (7/14), qnrS (7/14), aac (6')-Ib-cr (3/14), oqxA (12/14), and oqxB (11/14) with collective number reached five PAFQR genes per a single isolate. This finding agrees with the results reported by other authors who showed that imipenem sensitive A. baumannii isolates harbored certain resistance genes72, 73 and the same was exhibited by other members of gram-negative bacilli worldwide74. The occurrence of resistance genes in CIP-sensitive A. baumannii isolates represents a major problem as this could facilitates their horizontal transfer between A. baumannii and other members of gram-negative bacilli in hospitals.
Much earlier theory has given special affirmation for transferable antibiotic resistance among A. baumannii by plasmid-associated quinolone resistance determinant genes13, 59, 60. Depending on the sequence data analysis of PAFQR gene amplicons conducted in our study, it was demonstrated that oqxA and oqxB were identified previously in Klebsiella aerogenes strain NCTC9793 and Klebsiella pneumoniae, respectively with identity of 99.77%. While, the sequences of qnrS, and acc(6,)-ib-cr, were identified previously in A. baumannii with identity ranged of 98.28% and 97.98%, respectively. Our results agree that of Hamed et al. in the north Egypt area who reported that oqxAB was not detected in A. baumannii and E. coli while it was detected in Klebsiella spp25. This result suggests the possibility of acquisition of transferable antibiotic resistance genes that may be carried on plasmids by A. baumannii.
Our results showed the existence of several PAFQR genes and their co-occurrence in A. baumannii recovered isolates. The distribution of tested PAFQR genes gave 26 profiles for the 86 CIP-resistant- and 8 profiles for the 14 CIP-sensitive isolates (both harbor up to five genes). The storm of association due to suppleness and the non-limit capacity of A. baumannii for genome changes75, which may be caused by horizontal gene transfer76, the introduction of mobile genetic elements like plasmids which mediate new genes, integrative conjugative elements, and transposons60, 76. These properties give rise to PDR, XDR, and MDR gene cassettes77.
Conclusion
MDR, XDR, and PDR A. baumannii isolates are becoming prevalent in a number of hospitals, among the reasons behind this spread could be due to the empirical and un-rational use of antimicrobial agents which usually occur among outpatients in addition to improper infection control applied measures. It was observed that chromosomal mutations in the sequences of GyrA and ParC encoding genes as well as the acquisition of PAFQR encoding genes are molecular resistance mechanisms demonstrated among fluoroquinolones resistant A. baumannii isolates. It is advisable to monitor the antimicrobial resistance profiles of pathogens causing nosocomial infections and properly apply and update antibiotic stewardship in hospitals and for outpatients to control infectious diseases and prevent development of the microbial resistance to antimicrobial agents.
References
Lee, C.-R. et al. Biology of Acinetobacter baumannii: Pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front Cell Infect. Microbiol. 13, 7–55. https://doi.org/10.3389/fcimb.2017.00055 (2017).
Montefour, K. et al. Acinetobacter baumannii: An emerging multidrug-resistant pathogen in critical care. Crit. Care Nurse 28, 15–25. https://doi.org/10.4037/ccn2008.28.1.15 (2008).
Cerezales, M. et al. Mobile genetic elements harboring antibiotic resistance determinants in Acinetobacter baumannii isolates from bolivia. Front Microbiol. 11, 919–923. https://doi.org/10.3389/fmicb.2020.00919 (2020).
Mohammed, M. A. et al. Propranolol, chlorpromazine and diclofenac restore susceptibility of extensively drug-resistant (XDR)-Acinetobacter baumannii to fluoroquinolones. PLoS ONE 15, e0238195. https://doi.org/10.1371/journal.pone.0238195 (2020).
Tawfick, M. M. & El-Borhamy, M. I. PCR-RFLP-Based detection of mutations in the chromosomal fluoroquinolone targets gyrA and parC genes of Acinetobacter baumanii clinical isolates from a tertiary hospital in Cairo, Egypt. Am. J. Microbiol. Res. 5, 37–43. https://doi.org/10.12691/ajmr-5-2-3 (2017).
El Chakhtoura, N. G. et al. Therapies for multidrug resistant and extensively drug-resistant non-fermenting gram-negative bacteria causing nosocomial infections: A perilous journey toward “molecularly targeted” therapy. Exp. Rev. Anti Infect. Ther. https://doi.org/10.1080/14787210.2018.1425139 (2018).
Kakuta, N. et al. A novel mismatched PCR-restriction fragment length polymorphism assay for rapid detection of gyrA and parC mutations associated with fluoroquinolone resistance in Acinetobacter baumannii. Ann. Lab Med. 40, 27–32. https://doi.org/10.3343/alm.2020.40.1.27 (2020).
Martínez-Martínez, L., Pascual, A. & Jacoby, G. A. Quinolone resistance from a transferable plasmid. Lancet 351, 797–799. https://doi.org/10.3343/alm.2020.40.1.27 (1998).
Poirel, L., Cattoir, V. & Nordmann, P. Plasmid-mediated quinolone resistance; interactions between human, animal, and environmental ecologies. Front Microbiol. 3, 24. https://doi.org/10.3389/fmicb.2012.00024 (2012).
Hooper, D. C. & Jacoby, G. A. Topoisomerase inhibitors: Fluoroquinolone mechanisms of action and resistance. Cold Spring Harbor Perspect. Med. 6, a025320. https://doi.org/10.1101/cshperspect.a025320 (2016).
Robicsek, A. et al. Fluoroquinolone-modifying enzyme: A new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 12, 83–88. https://doi.org/10.1038/nm1347 (2006).
Yamane, K. et al. New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. Antimicrob Agents Chemother. 51, 3354–3360. https://doi.org/10.1128/AAC.00339-07 (2007).
Yang, H. et al. Detection of the plasmid-mediated quinolone resistance determinants in clinical isolates of Acinetobacter baumannii in China. J. Chemother. 28, 443–445. https://doi.org/10.1179/1973947815Y.0000000017 (2016).
Touati, A. et al. First report of qnrB-producing Enterobacter cloacae and qnrA-producing Acinetobacter baumannii recovered from Algerian hospitals. Diagn. Microbiol. Infect. Dis. 60, 287–290. https://doi.org/10.1016/j.diagmicrobio.2007.10.002 (2008).
Jiang, X. et al. Emergence of plasmid-mediated quinolone resistance genes in clinical isolates of Acinetobacter baumannii and Pseudomonas aeruginosa in Henan China. Diagn. Microbiol. Infect. Dis. 79, 381–383. https://doi.org/10.1016/j.diagmicrobio.2014.03.025 (2014).
Koneman, E. W. & Allen, S. Koneman. Diagnostico Microbiologico/ Microbiological diagnosis: Texto Y Atlas En Color/Text and Color Atlas. (Ed. médica panamericana, (2008).
Gerner-Smidt, P., Tjernberg, I. & Ursing, J. Reliability of phenotypic tests for identification of Acinetobacter species. Clin. Microbiol. 29, 277–282. https://doi.org/10.1128/JCM.29.2.277-282.1991 (1991).
Turton, J. F. et al. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. Clin. Microbiol. 44, 2974–2976. https://doi.org/10.1128/JCM.01021-06 (2006).
CLSI. Performance standards for antimicrobial susceptibility testing. Twentieth informational supplement, document M100s-S27. Clinical and Laboratory Standards Institute. Wayne, PA. USA (2017).
Falagas, M. E. & Karageorgopoulos, D. E. Pandrug resistance (PDR), extensive drug resistance (XDR), and multidrug resistance (MDR) among Gram-negative bacilli: Need for international harmonization in terminology. Clin. Infect. Dis. 46, 1121–1122. https://doi.org/10.1086/528867 (2008).
Versalovic, J., Koeuth, T. & Lupski, R. Distribution of repetitive DNA sequences in eubacteria and application to finerpriting of bacterial enomes. Nucleic Acids Res. 19, 6823–6831. https://doi.org/10.1093/nar/19.24.6823 (1991).
Tsai, H.-C. et al. Acinetobacter baumannii and methicillin-resistant Staphylococcus aureus in long-term care facilities in eastern Taiwan. Tzu-Chi Med. J. 31, 222. https://doi.org/10.4103/tcmj.tcmj_136_18 (2019).
Hamed, S. M. et al. Multiple mechanisms contributing to ciprofloxacin resistance among Gram negative bacteria causing infections to cancer patients. Sci. Rep. 8, 1–10. https://doi.org/10.1038/s41598-018-30756-4 (2018).
Lee, J. K., Lee, Y. S., Park, Y. K. & Kim, B. S. Mutations in the gyrA and parC genes in ciprofloxacin-resistant clinical isolates of Acinetobacter baumannii in Korea. Microbiol. Immunol. 49, 647–653. https://doi.org/10.1111/j.1348-0421.2005.tb03643.x (2005).
Hamed, S. M. et al. Plasmid-mediated quinolone resistance in gram-negative pathogens isolated from Cancer patients in Egypt. Microbiol. Drug Resist. 24, 1316–1325. https://doi.org/10.1089/mdr.2017.0354 (2018).
Gant, V. A. et al. Three novel highly charged copper-based biocides: Safety and efficacy against healthcare-associated organisms. J. Antimicrob. Chemother. 60, 294–299. https://doi.org/10.1093/jac/dkm201 (2007).
Eichner, A. et al. Novel photodynamic coating reduces the bioburden on near-patient surfaces thereby reducing the risk for onward pathogen transmission: A field study in two hospitals. Hosp. Infect. 104, 85–91. https://doi.org/10.1016/j.jhin.2019.07.016 (2020).
Verma, P., Maurya, P., Tiwari, M. & Tiwari, V. In-silico interaction studies suggest RND efflux pump mediates polymyxin resistance in Acinetobacter baumannii. J. Biomol. Struct. Dyn. 37, 95–103. https://doi.org/10.1080/07391102.2017.1418680 (2019).
Asif, M., Alvi, I. A. & Rehman, S. U. In sight into Acinetobacter baumannii: pathogenesis, global resistance, mechanisms of resistance, treatment options, and alternative modalities. Infect. Drug Resist. 11, 1249. https://doi.org/10.2147/IDR.S166750 (2018).
Abd El-Baky, R. M. et al. Antimicrobial resistance pattern and molecular epidemiology of ESBL and MBL producing Acinetobacter baumannii isolated from hospitals in Minia. Egypt. Alex Med. 56, 4–13. https://doi.org/10.1080/20905068.2019.1707350 (2020).
Dolzani, L. et al. Identification of Acinetobacter isolates in the A. calcoaceticus-A. baumannii complex by restriction analysis of the 16S–23S rRNA intergenic-spacer sequences. J. Clin. Microbiol. 33, 1108–1113. https://doi.org/10.1128/JCM.33.5.1108-1113.1995 (1995).
Chaturvedi, R., Chandra, P. & Mittal, V. Biofilm formation by Acinetobacter spp. in association with antibiotic resistance in clinical samples obtained from tertiary care hospital. Res. J. Pharm. Technol. 12, 3737–3742. https://doi.org/10.5958/0974-360X.2019.00620.6 (2019).
Sun, R.-X. et al. Morbidity and mortality risk factors in emergency department patients with Acinetobacter baumannii bacteremia. World J. Emerg. Med. 11, 164. https://doi.org/10.5847/wjem.j.1920-8642.2020.03.006 (2020).
Piano, S. et al. Epidemiology and effects of bacterial infections in patients with cirrhosis worldwide. Gastroenterology 156, 1368–1380. https://doi.org/10.1053/j.gastro.2018.12.005 (2019).
Abdulzahra, A. T., Khalil, M. A. & Elkhatib, W. F. First report of colistin resistance among carbapenem-resistant Acinetobacter baumannii isolates recovered from hospitalized patients in Egypt. New Microbes New Infect. 26, 53–58. https://doi.org/10.1016/j.nmni.2018.08.007 (2018).
López-Durán, P. A. et al. Nosocomial transmission of extensively drug resistant Acinetobacter baumannii strains in a tertiary level hospital. PLoS ONE 15, e0231829. https://doi.org/10.1371/journal.pone.0231829 (2020).
Corrêa, R. C., Heleno, S. A., Alves, M. J. & Ferreira, I. C. Bacterial resistance: Antibiotics of last generation used in clinical practice and the arise of natural products as new therapeutic alternatives. Curr. Pharm. Des. 26, 815–837. https://doi.org/10.2174/1381612826666200224105153 (2020).
Isler, B., Doi, Y., Bonomo, R. A. & Paterson, D. L. New treatment options against carbapenem-resistant Acinetobacter baumannii infections. Antimicrob. Agents Chemother. 63, e01110-01118. https://doi.org/10.1128/AAC.01110-18 (2019).
Karakonstantis, S., Kritsotakis, E. I. & Gikas, A. Pandrug-resistant Gram-negative bacteria: a systematic review of current epidemiology, prognosis and treatment options. J. Antimicrob. Chemother. 75, 271–282. https://doi.org/10.1093/jac/dkz401 (2020).
Dekic, S., Hrenovic, J., Durn, G. & Venter, C. Survival of extensively-and pandrug-resistant isolates of Acinetobacter baumannii in soils. Appl. Soil. Eco. 147, 103396. https://doi.org/10.1016/j.apsoil.2019.103396 (2020).
Aydemir, H. et al. Risk factors and clinical responses of pneumonia patients with colistin-resistant Acinetobacter baumannii-calcoaceticus. World J. Clin. Cases 7, 1111–1121. https://doi.org/10.12998/wjcc.v7.i10.1111 (2019).
Yuan, X., Liu, T. & Di, W. Epidemiology, susceptibility, and risk factors for acquisition of MDR/XDR gram-negative bacteria among kidney transplant recipients with urinary tract infections. Infect. Drug. Resist. 11, 707. https://doi.org/10.2147/IDR.S163979 (2018).
Baccolini, V. et al. Effectiveness over time of a multimodal intervention to improve compliance with standard hygiene precautions in an intensive care unit of a large teaching hospital. Antimicrob. Resist. Infect. Control 8, 92. https://doi.org/10.1186/s13756-019-0544-0 (2019).
Hornsey, M. & Wareham, D. W. Effects of in vivo emergent tigecycline resistance on the pathogenic potential of Acinetobacter baumannii. Sci. Rep. 8, 1–8. https://doi.org/10.1038/s41598-018-22549-6 (2018).
Falah, F., Shokoohizadeh, L. & Adabi, M. Molecular identification and genotyping of Acinetobacter baumannii isolated from burn patients by PCR and ERIC-PCR. Scars Burn Heal 5, 2059513119831369. https://doi.org/10.1177/2059513119831369 (2019).
Versalovic, J., Schneider, M., De Bruijn, F. & Lupski, J. R. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Meth. Molec. Cell Biol. 5, 25–40 (1994).
Biswas, D., Tiwari, M. & Tiwari, V. Molecular mechanism of antimicrobial activity of chlorhexidine against carbapenem-resistant Acinetobacter baumannii. PLoS ONE https://doi.org/10.1371/journal.pone.0224107 (2019).
Attia, N. M. & ElBaradei, A. Fluoroquinolone resistance conferred by gyrA, parC mutations, and AbaQ efflux pump among Acinetobacter baumannii clinical isolates causing ventilator-associated pneumonia. Acta Microbiol. Immunol. Hung 67, 234–238. https://doi.org/10.1556/030.66.2019.040 (2019).
Santhosh, K. S. et al. Multiple antimicrobial resistance and novel point mutation in fluoroquinolone-resistant Escherichia coli isolates from Mangalore India. Microbiol. Drug Resist. 23, 994–1001. https://doi.org/10.1089/mdr.2016.0142 (2017).
Sun, C., Hao, J., Dou, M. & Gong, Y. Mutant prevention concentrations of levofloxacin, pazufloxacin and ciprofloxacin for A. baumannii and mutations in gyrA and parC genes. Antibiotic 68, 313–317. https://doi.org/10.1038/ja.2014.150 (2015).
Mahapatra, A. et al. Ciprofloxacin-resistant Gram-negative isolates from a tertiary care hospital in Eastern India with novel gyrA and parC gene mutations. Med. Arm Forc. India. https://doi.org/10.1016/j.mjafi.2019.10.002 (2020).
Abdelkreem, R. H., Yousuf, A. M., Elmekki, M. A. & Elhassan, M. M. DNA gyrase and topoisomerase IV mutations and their effect on quinolones resistant proteus mirabilis among UTIs patients. Pak J. Med. Sci. 36, 1234–1240. https://doi.org/10.12669/pjms.36.6.2207 (2020).
Lindbäck, E., Rahman, M., Jalal, S. & Wretlind, B. Mutations in gyrA, gyrB, parC, and parE in quinolone-resistant strains of Neisseria gonorrhoeae. APMIS 110, 651–657. https://doi.org/10.1034/j.1600-0463.2002.1100909.x (2002).
Tokue, Y. et al. Detection of novel mutations in the gyrA gene of Staphylococcus aureus by nonradioisotopic single-strand conformation polymorphism analysis and direct DNA sequencing. Antimicrob. Agents Chemother. 38, 428–431. https://doi.org/10.1128/aac.38.3.428 (1994).
Takenouchi, T. et al. Incidence of various gyrA mutants in 451 Staphylococcus aureus strains isolated in Japan and their susceptibilities to 10 fluoroquinolones. Antimicrob. Agents Chemother. 39, 1414–1418. https://doi.org/10.1128/aac.39.7.1414 (1995).
Safari, M. et al. Sequence-based detection of first-line and second-line drugs resistance-associated mutations in Mycobacterium tuberculosis isolates in Isfahan. Iran. Infect. Genet. Evol. https://doi.org/10.1016/j.meegid.2020.104468 (2020).
Felde, O. et al. Development of molecular biological tools for the rapid determination of antibiotic susceptibility of Mycoplasma hyopneumoniae isolates. Vet Microbiol. 10, 8697. https://doi.org/10.1016/j.vetmic.2020.108697 (2020).
Hongbo, L., Zhen, S., Xiaoman, X. & Shengqi, L. Mutations in the quinolone resistance determining region in isogenic mutant Acinetobacter baumannii strains. Afr. J. Microbiol. Res. 7, 3559–3562. https://doi.org/10.5897/AJMR2013.5932 (2013).
Saranathan, R. et al. Multiple drug resistant carbapenemases producing Acinetobacter baumannii isolates harbours multiple R-plasmids. Indian J. Med. Res. 140, 262–270 (2014).
Salto, I. P. et al. Comparative genomic analysis of Acinetobacter spp. plasmids originating from clinical settings and environmental habitats. Sci. Rep. 8, 1–12. https://doi.org/10.1038/s41598-018-26180-3 (2018).
Siddiqui, M. T. et al. Plasmid-mediated ampicillin, quinolone, and heavy metal co-resistance among ESBL-producing isolates from the Yamuna River, New Delhi India. Antibiotic 9, 826. https://doi.org/10.3390/antibiotics9110826 (2020).
Yanat, B., Rodríguez-Martínez, J.-M. & Touati, A. Plasmid-mediated quinolone resistance in Enterobacteriaceae: A systematic review with a focus on Mediterranean countries. Eur. J. Clin. Microbiol. Infect. Dis. 36, 421–435. https://doi.org/10.1007/s10096-016-2847-x (2017).
Jacoby, G. A. Mechanisms of resistance to quinolones. Clin. Infect. Dis. 41, S120–S126. https://doi.org/10.1086/428052 (2005).
Ali, S. A., Hassan, R. M., Khairat, S. M. & Salama, A. M. Plasmid-Mediated Quinolone Resistance Genes qnrA, qnrB, qnrC, qnrD, qnrS and aac (6’)-Ib in Pseudomonas aeruginosa and Acinetobacter baumannii. Microbiol. Res. J. Int. https://doi.org/10.9734/MRJI/2018/44295 (2018).
Piekarska, K. et al. Co-existence of plasmid-mediated quinolone resistance determinants and mutations in gyrA and parC among fluoroquinolone-resistant clin Enterobacteriaceae isolated in a tertiary hospital in Warsaw Poland. Int. J. Antimicrob. Agents 45, 238–243. https://doi.org/10.1016/j.ijantimicag.2014.09.019 (2015).
Gogry, F. A., Siddiqui, M. T. & Haq, Q. M. R. Emergence of mcr-1 conferred colistin resistance among bacterial isolates from urban sewage water in India. Environ. Sci. Poll. Res. Int. 26, 33715–33717. https://doi.org/10.1007/s11356-019-06561-5 (2019).
Vetting, M. W. et al. Mechanistic and structural analysis of aminoglycoside N-acetyltransferase AAC (6′)-Ib and its bifunctional, fluoroquinolone-active AAC (6′)-Ib-cr variant. Biochemistry 47, 9825–9835. https://doi.org/10.1021/bi800664x (2008).
Mirnejad, R. et al. Evaluation of Polymyxin B susceptibility profile and detection of drug resistance genes among Acinetobacter baumannii Clinical Isolates in Tehran, Iran during 2015–2016. Mediterr. J. Hematol. Infect. Dis. 10, e2018044. https://doi.org/10.4084/MJHID.2018.044 (2018).
Choori, M., Eftekhar, F. & Saniee, P. Distribution of plasmid-mediated quinolone resistance, integrons and AdeABC efflux pump genes in nosocomial isolates of Acinetobacter baumannii. J. Krish Instit. Med. Sci. 8, 22–30 (2019).
Kim, H. B. et al. oqxAB encoding a multidrug efflux pump in human clinical isolates of Enterobacteriaceae. Antimicrob. Agents Chemother. 53, 3582–3584. https://doi.org/10.1128/AAC.01574-08 (2009).
Miškinytė, M. et al. High incidence of plasmid-mediated quinolone resistance (PMQR) genes among antibiotic-resistant Gram-negative bacteria from patients of the Lithuanian National Cancer Center. Infect. Dis. 51, 471–474. https://doi.org/10.1080/23744235.2019.1591637 (2019).
Gomaa, F.A.-Z.M., Tawakol, W. M. & Abo El-Azm, F. I. Phenotypic and genotypic detection of some antimicrobial resistance mechanisms among multidrug-resistant Acinetobacter baumannii isolated from immunocompromised patients in Egypt. Egypt J. Med. Microbiol. 38, 1–14 (2014).
Daef, E. A. et al. Evaluation of different phenotypic assays for the detection of metallo-β-lactamase production in carbapenem susceptible and resistant Acinetobacter baumannii isolates. J. Am. Sci. 8, 292–299 (2012).
Goyal, R. & Sarma, S. Existence of Metallo beta lactamases in Carbapenem susceptible gram negative bacilli: A cause for concern. J. Clin. Diag. Res. 4, 2679–2684 (2010).
Gordon, N. C. & Wareham, D. W. Multidrug-resistant Acinetobacter baumannii: mechanisms of virulence and resistance. Int. J. Antimicrob. Agents 35, 219–226. https://doi.org/10.1016/j.ijantimicag.2009.10.024 (2010).
Arber, W. Genetic variation: Molecular mechanisms and impact on microbial evolution. FEMS Microbiol. Rev. 24, 1–7. https://doi.org/10.1111/j.1574-6976.2000.tb00529.x (2000).
Weldhagen, G. F. Integrons and β-lactamases—a novel perspective on resistance. Int. J. Antimicrob. Agents 23, 556–562. https://doi.org/10.1016/j.ijantimicag.2004.03.007 (2004).
Acknowledgements
The authors are grateful to the staffs at Medical Research Center, Faculty of Medicine Assiut University, and Al Azhar University hospitals for providing A. buamannii isolates. More grateful is extended to Prof. Enas A Deaf, and Prof. Nahla M Elsherbiny; Department of Microbiology and Immunology, Faculty of Medicine, Assiut University for the facility and help provided by them during this work. All my appreciation to Dr. Mamdouh A. Younes; Department of Microbiology and Immunology, Faculty of Pharmacy, Al-Azhar University, Assiut branch for his help during the study. Also, more gratitude is extended to Dr. Mohammed Eissa, INRS, University of Quebec, Canada, and Dr. Samira M. Hamed, Department of Microbiology and Immunology, Faculty of Pharmacy, October University for Modern Sciences and Arts for their support and help during this study.
Funding
This research received no internal and external funding.
Author information
Authors and Affiliations
Contributions
M.A.M.: Conceptualization, Data curation, Investigation, Formal analysis, Methodology, Writing—original draft. M.T.A.S.: Conceptualization, Formal analysis, Methodology, Writing—review & editing. B.E.A.: Conceptualization, Data curation, Investigation, Methodology, review—original draft. K.M.A.: Conceptualization, Formal analysis, Investigation, Methodology, and Supervision. M.M.A.: Conceptualization, Formal analysis, Investigation, Methodology, Supervision, Writing—review & editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mohammed, M.A., Salim, M.T.A., Anwer, B.E. et al. Impact of target site mutations and plasmid associated resistance genes acquisition on resistance of Acinetobacter baumannii to fluoroquinolones. Sci Rep 11, 20136 (2021). https://doi.org/10.1038/s41598-021-99230-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-99230-y
- Springer Nature Limited