Abstract
Coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerges to scientific research and monitoring of wastewaters to predict the spread of the virus in the community. Our study investigated the COVID-19 disease in Bratislava, based on wastewater monitoring from September 2020 until March 2021. Samples were analyzed from two wastewater treatment plants of the city with reaching 0.6 million monitored inhabitants. Obtained results from the wastewater analysis suggest significant statistical dependence. High correlations between the number of viral particles in wastewater and the number of reported positive nasopharyngeal RT-qPCR tests of infected individuals with a time lag of 2 weeks/12 days (R2 = 83.78%/R2 = 52.65%) as well as with a reported number of death cases with a time lag of 4 weeks/27 days (R2 = 83.21%/R2 = 61.89%) was observed. The obtained results and subsequent mathematical modeling will serve in the future as an early warning system for the occurrence of a local site of infection and, at the same time, predict the load on the health system up to two weeks in advance.
Similar content being viewed by others
Introduction
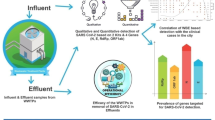
A thorough understanding of the current pandemic of COVID-19 is for public health officials a critical and ongoing challenge1. Currently, there are several epidemiological tools to gain control, however all of them have limitations. Rapid diagnostics tests are not accurate enough, the capacity of RT-qPCR testing may be insufficient, tracing contacts is based on personnel capacities and primarily symptomatic individuals are reported1,2. Long-term international experience with systematic monitoring of illegal drugs and their metabolites proposes that wastewater-based epidemiology (WBE) can be a highly effective way of surveillance for the presence of specific pathogens in communities3,4,5,6 (Fig. 1). Furthermore, it can be used to monitor the increasing or decreasing trend of the pathogen and its transmission1,2. The occurrence of hotspots caused by viruses may induce problems, especially in densely populated areas. Classical epidemiology lacks any prediction of highly infectious locations and is based mainly on clinical symptoms of infected individuals. Therefore in the case of COVID-19, where symptoms are delayed and many asymptomatic patients are observed, it has difficulties recognizing the acute onset of the disease2. SARS-CoV-2 or Severe Acute Respiratory Syndrome Coronavirus 2 causing respiratory illness COVID-19 is infectious diseases that resulted in 216 303 376 diagnosed cases and 4 498 451 deaths as of 30th August 2021 according to WHO7. Individuals infected by the virus exhibit various symptoms, but most often shortness of breath, dry cough and fever. The symptoms onset is usually within 2–14 days after exposure to the virus8,9. Nevertheless, around 45% of infected individuals show no symptoms of being asymptomatic throughout the course of the disease10,11.
At the beginning of 2020, several studies pointed out that viral particles of SARS-CoV-2 may be shed to the feces of infected individuals and therefore occur in the sewage system12,13,14. The assumption resides in the ability of SARS-CoV-2 to bind to angiotensin-converting enzyme 2 receptor (ACE2) that is abundant in the small intestine15,16. According to studies, viral particles are present in the stool early after infection and remain present during disease (9–16 days post symptom onset). Therefore they can be detected similarly as nasopharyngeal swabs in case of their negativity17,18. Based on these observations, WBE of SARS-CoV-2 may offer valuable additional and early information of infection tendency in a community19.
Wastewater-based monitoring of COVID-19 offers several advantages. Compared to clinical testing, it is a noninvasive cost-effective way of investigating virus transmission dynamics in entire communities. Moreover, there is a possibility of monitoring populations without access to standard healthcare systems. On the contrary, the downsides of the methods are a difficult analytical measurement of viral RNA, inconsistency in ongoing analyses with high starting costs20. Monitoring of SARS-CoV-2 can be further hampered by high portions of ballast waters and the chemical or biological composition of wastewater which may differ within the monitored region.
Our study describes the monitoring of RNA SARS-CoV-2 in wastewaters and defines a mathematical model of possible relations between RNA SARS-CoV-2 concentration in wastewater and subsequent increase of positive and death cases. Long-term monitoring was carried out in Bratislava, with an estimated population of 0.6 million inhabitants from September 2020 until March 2021.
Materials and methods
Characterization of investigated locations
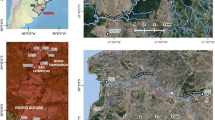
Bratislava is a capital and the most populated city in the Slovak Republic, with approximately 600,000 inhabitants (Fig. 2). Petrzalka, with its satellites has a population of about 125,000 residents. It is the largest suburb of Bratislava (district Bratislava V) with the highest population density in Slovakia as well as all of Central Europe. It is located next to the city center on the right bank of the Danube River.
Geographical map of Slovakia and Bratislava with the districts. Green districts collect sewage to wastewater treatment plant (WWTP) Bratislava—Centrum (Vrakuna), represent 450,000 inhabitants. Red districts are areas containing sewage to WWTP Bratislava-Petrzalka representing 125,000 inhabitants. Map of Slovakia (https://sk.wikipedia.org/wiki/S%C3%BAbor:Slovakia_-_outline_map.svg) is licensed Creative Commons Attribution-ShareAlike 3.0 license. The license terms can be found on the following link: https://creativecommons.org/licenses/by-sa/3.0/. Map of Bratislava (https://sk.m.wikipedia.org/wiki/S%C3%BAbor:Bratislava_boroughs_outline_map.svg) is licensed Creative Commons Attribution-ShareAlike 4.0 license. The license terms can be found on the following link: https://creativecommons.org/licenses/by-sa/4.0/deed.cs (Edited by Tamas M, Inkscape, 1.0beta1, https://inkscape.org/).
Characterization of the wastewater treatment plants (WWTPs)
This study focused on the incidence of selected SARS-CoV-2 genes in two different Slovak (Bratislava) WWTPs. The first one is located in Bratislava—Centrum that treats almost all wastewater from the significant part of Bratislava. The second one is in Bratislava—Petrzalka that treats wastewater from Petrzalka (Table 1). WWTPs use a mechanical and biological stage (stage nitrification only); the produced sludge is digested and produced biogas energy is recovered.
Sampling
Automatic sampler devices were used to collect raw wastewater samples at the inflow point of WWTPs. Monitoring of wastewater was carried out at Bratislava—Centrum and Bratislava—Petrzalka from September 2020 until March 2021. During the sampling campaign more than 50 samples of influent were taken (Tables 1, 2).
Samples with a volume of 50 ml were taken every 15 min for a period of 24 h to obtain a daily representative sample (in total 4.8 l). The collected specimen was frozen within 2 h at − 20 °C and kept at this temperature until processing. This procedure follows a protocol established in several previous studies to prevent the material from biodegradation21.
Sample processing
50 ml of wastewater was centrifuged at 4700 g for 30 min, and then 45 ml of clear supernatant was subjected to ultracentrifugation 125,893 g for 90 min. After ultracentrifugation, the pellet was resuspended in 1 ml of TRI reagent. RNA was isolated using Direct-zol RNA Miniprep (Zymo Research) according to the manufacturer’s instructions. Each sample was eluted in 100 µl of RNase-free water and RNA quantity was measured using NanoDrop ND-2000 (Nanodrop Inc.). A maximum of 8 µg of RNA per sample was concentrated by vacuum evaporation using Eppendorf Concentrator plus (Eppendorf).
SARS-CoV-2 detection using quantitative RT-qPCR
The concentrated sample was resuspended in 11 µl of RNase-free water. A RevertAid First Strand cDNA synthesis kit (ThermoFisher Scientific) in a total volume of 20 μl was used for reverse transcription according to the manufacturer's instructions. 2 µl of the reverse transcription reaction product was analyzed in one RT-qPCR reaction using PerfeCTa qPCR FastMix II, low ROX (VWR) and specific primers for E, RdRP (described by WHO) and ORF1ab, S (described by Sigma Aldrich) genes (Supplementary Table S1). Initial denaturation was set to 10 min at 95 °C followed by 45 cycles of denaturation (95 °C, 30 s), annealing (59 °C, 30 s) and extension (72 °C, 30 s) using Stratagene Mx3005P (Agilent). Copies were estimated based on standard curves using synthetic SARS-CoV-2 RNA Control 1 (Australia/VIC01/2020, Genbank ID: MT007544.1).
RT-qPCR product confirmation by Sanger sequencing
RT-qPCR products of each measured gene were further confirmed by Sanger sequencing. Out of all samples 8 were chosen for the confirmation. Sanger sequencing of the amplicons was carried by Microsynth Austria GmbH (Vienna, Austria). Quality control of sequences were assessed and confirmed in the BLAST standard database of nucleotide collection for coronavirus detection. Only highly similar sequences were reported. Reports from sequencing and BLAST results are accessible in Supplementary_material_2.
Calculation of viral particles in sewage and mathematical modeling
Regression models (simple linear, Double Squared Root and Square root-Y logarithmic-X) have been used to describe the studied relations between the obtained data from wastewater and reported data related to the COVID-19 situation. The time-series analysis was used to compare the dependence between the wastewater time series and various time lags of the positive RT-qPCR tests (and Death cases) time series. To determine how well the time lags match up with the wastewater time series and in particular, at what point the best match (dependence with the highest R2) occurs, the cross-correlation function was applied. A generalized additive model (GAM) was used to illustrate the smoothed curves of relevant time series. The one-way ANOVA described the differences between the means of the positive RT-qPCR tests in particular days of the week based on the Studentized range statistic, Tukey's 'Honest Significant Difference' method.
The method's detection limit was calculated as the number of monitored inhabitants divided by minimal positive cases reported at the date with positive RT-qPCR wastewater analysis for SARS-CoV-2.
All statistical and mathematical operations were performed in the R environment and software program StatGraphics22.
Results and discussion
Feces are the main contribution of SARS-CoV-2 presence in sewage, thus selected target genes were likewise detected by several research groups in feces or from rectal swabs15,18,23. Detection of several genes is vital as the RNA virus in wastewater is exposed to many environmental factors. Therefore, RNA may be strongly disintegrated resulting in poor amplification and variable detection of individual viral particles. We focused on detecting four genes ORF1ab, S, E and RdRp gene capturing different regions of the virus24. Similar target genes (S, RdRP, ORF1ab) were also selected by La Rossa et al.25. On the contrary, other researchers were investigating the presence of 3 different regions of nucleocapsid (N) gene in wastewater26. Currently, there is no consensus of targets recommended for detecting SARS-CoV-2 in sewage.
In our study 29 out of 52 analyzed wastewater samples (56%) were found to be positive. Table 2 displays CT values of detected genes in both WWTPs at given dates. First positive results were obtained on 29th September with detected S gene at both WWTPs and ORF1ab gene at WWTP Bratislava-Petrzalka. In Bratislava—Centrum lowest CT detected was 31.8, whereas in Bratislava—Petrzalka, it was 32.7 on the same date. At least one of the targets was detected since this date at chosen WWTPs. The most often detected target was the ORF1ab gene with positivity of 33% (22 out of 66) from all positively detected targets.
Further, amplicons generated by RT-qPCR from 8 samples were confirmed by Sanger sequencing. This was done to prevent false negative results often obtained by RT-qPCR analysis as wastewater contains various RNA fragments and cross-reacting molecules27. 7 out of 8 passed quality control check and the amplicon sequences were aligned to coronavirus strain with 5 sequences confirming SARS-CoV-2 as first hits in the BLAST database (Supplementary table S3, Supplementary_material_2).
According to calibration curves, CTs presented in Table 2 were recalculated to the number of viral particles per milliliter. To calculate the daily loads of SARS-CoV-2 in wastewater, viral particles per ml were then multiplied by daily influents measured at the WWTPs (Supplementary Table S2). Then reported numbers of positive RT-qPCR tests and reported numbers of death cases were compared to the number of viral particles in wastewater in Bratislava (Fig. 3).
By interpolating the measured viral particles in the wastewater, we obtained a model useful for comparing the time series of positive RT-qPCR tests, death cases and viral particles (Fig. 3). Since the graph reveals apparent time shifts, we focused on the time lags between particular time series in our analysis.
The following Table 3 describes the dependencies between the time lags of reported positive RT-qPCR tests or death cases and the number of viral particles found on individual days in the wastewater of Bratislava.
It can be seen that the values of R2 for dependence between viral particles in wastewater and reported positive RT-qPCR tests and death cases (Table 3, Supplementary Fig. S2, S3) are better for the weekly time series than for the daily time series. This led us to focus our analysis on weekly time series. This decision is supported by the results of the ANOVA test where the significant difference among the means of the number of positive RT-qPCR tests in the particular days of the week was confirmed (p-value = 0.0054, F-value = 3.178, DF = 6, Supplementary Fig. S1). Therefore, we decided to use the time series of cumulative data with a 7 days period. The weekly time series of measured viral particles in wastewater reported numbers of positive RT-qPCR tests and reported numbers of death cases without/with the appropriate time lags are depicted in supplementary Fig. S4.
By analyzing the dependence between the weekly time series of measured viral particles in the wastewater and reported numbers of positive RT-qPCR tests with a time lag of 2 weeks, the models for estimation of the number of positive RT-qPCR tests were obtained. The results of fitting a linear (R2 = 81.72%, p-value < 0.0001, F-value = 116.2, DF = 26, Supplementary Fig. S5) and double square root (R2 = 83.78%, p-value < 0.0001, F-value = 134.3, DF = 26, Supplementary Fig. S6) models describe the relationship between the weekly time series of measured viral particles in the wastewater and reported numbers of positive RT-qPCR tests with the time lag of 2 weeks. The equations of the fitted models are:
Linear:
Double square root:
The models allow estimating the number of positive RT-qPCR tests with model fitting more than 80% (Figure S7).
By analyzing the dependence between the weekly time series of measured viral particles in the wastewater and reported numbers of death cases with a time lag of 4 weeks, the models for estimation of the number of death cases were also obtained. The results of fitting a linear (R2 = 48.06%, p-value < 0.0001, F-value = 24.06, DF = 26, Supplementary Fig. S8) and square root-Y logarithmic-X (R2 = 83.21%, p-value < 0.0001, F-value = 128.83, DF = 26, Supplementary Fig. S9) models describe the relationship between the weekly time series of measured viral particles in the wastewater and reported numbers of death cases with the time lag of 4 weeks. The values of R2 show that the non-linear model is significantly better than the linear. The equations of the fitted models are:
Linear:
Square root-Y logarithmic-X:
The non-linear model allows estimating the number of death cases with model fitting more than 80% (Supplementary Fig. S10).
Further, we looked at the detection limit of wastewater monitoring related to the number of positive RT-qPCR cases within a 12 days shift. Individually, the detection limit of WWTP Bratislava—Vrakuna is 1 RT-qPCR positive case per 25,000 people on 15/3/2021, while Bratislava—Petrzalka reached 1 per 4,808 on 19/1/2021. The results are also limited by reporting the positive RT-qPCR cases as separate data for each city district has been available since 10/11/2020. For Bratislava the detection limit was 1 per 8,099 on 29/9/2020 when both WWTPs were combined. It is important to stress that the results are related and strongly biased to reported RT-qPCR positive cases which vary between the countries, their testing capacities and contact tracing. In comparison, the study by Ahmed et al., could not detect SARS-CoV-2 in wastewater until 100 reported cases per 100,000 people. On the contrary, Medema et al., was capable of detecting SARS-CoV-2 in the wastewater before the first confirmed results in the monitored area28,29. Another reason for such contrasting results is probably the different quality of the wastewater and various methodological approach influencing the detection limit. Each sewer has unique properties such as temperature, pH, presence of chemical substances and biological composition. All of these factors play a role in the degradation of viral particles. Therefore investigation of these properties is crucial and each methodological approach to detect SARS-CoV-2 from wastewater is unique for a given sewer system. However, some unification of the methodology would be vital for future comparison of the results.
Until now there was no confirmed case of infection from wastewater thus oral-fecal route is unlikely. The reason may be the inactivation of SARS-CoV-2 by gastrointestinal fluids or an unfavorable wastewater environment16,30,31. The amount of the virus shed to the feces may vary in time as well as between the patients23. Once the virus or its particles enter the sewage system, it is diluted by other types of water (industrial, rainfalls, water from snow melting, etc.) and is exposed to various physical and chemical factors. So far, viral RNA appears to be stable until reaching the primary settling tank of WWTP. Compared to non-enveloped viruses, SARS-CoV-2 has an affinity to wastewater solids, so a portion of the virus is probably sorbed on sewage walls and later to primary sludge30,32. On the contrary, wastewater surveillance offers several benefits once we have a working monitoring system. In September 2020, Larsen and Wigginton published in Nature Biotechnology that theoretically, wastewater surveillance can be in 7 days lead before rapid diagnostics tests and 13–15 days before delayed diagnostic tests1,33. This is in agreement with our observations where results from wastewater dated to the day of sampling were in 14 days (2 weeks) lead before standard clinical RT-qPCR testing. If we add 1–2 days of sample pretreatment and analysis we are reaching for 12–13 days advance before reported positive RT-qPCR tests. Novelty in the presented study is that a similar correlation can be found with deaths reported in 28 days (4 weeks) after the detected increase of viral particles in the wastewater. Although we can not specify the number of infected persons, observation of trends in near real-time can help us understand community transmission. In communities with low capacity of clinical testing and delays of diagnostics wastewater surveillance can be a temporary solution. Moreover, as it is relatively cost-effective and less invasive, wastewater monitoring can be used in low-income countries34,35. However in communities with working traditional testing, data obtained from wastewater are only additional information in controlling pandemics. They can be used to check the reliability of the introduction of novel technologies or protocols.
For successful monitoring of SARS-CoV-2 in the wastewater on a larger scale, public authorities must realize the importance of possible scalability of the method36. National agencies should show interest and provide financial and material support for research teams and water companies in the form of grants. Monitoring of COVID-19 and its mutations will require modification of the methods when compared to the monitoring of drugs or metabolites36. Optimistic presumption for successful cooperation is the fact that across Europe there is a working consortium of institutes and universities focusing on monitoring illegal drugs in the wastewaters37,38. European Commission at the beginning of March created the HERA incubator. One of the focuses is the systematic surveillance of the wastewaters, including genetic sequencing at WWTPs with a connected population of over 150,000 individuals39.
In the presented study, we detected RNA SARS-CoV-2 in wastewaters and displayed mathematical correlations between tested wastewater samples, positive RT-qPCR tests and death cases in Bratislava, Slovakia. The obtained results and subsequent mathematical modeling will be able to serve in the future as an early warning system for the occurrence of a local site of infection and at the same time will allow to predict the load on the health system up to two weeks in advance. Because each wastewater has its own characteristics (pH, temperature, (bio)chemical composition, etc.), it is necessary to approach this in other monitored localities when taking and processing samples, evaluating the results and creating the appropriate mathematical model.
References
Larsen, D. A. & Wigginton, K. R. Tracking COVID-19 with wastewater. Nat. Biotechnol. 38, 1151–1153 (2020).
Xagoraraki, I. & O’Brien, E. Wastewater-based epidemiology for early detection of viral outbreaks. In Women in Water Quality: Investigations by Prominent Female Engineers (ed. O’Bannon, D. J.) 75–97 (Springer International Publishing, 2020). https://doi.org/10.1007/978-3-030-17819-2_5.
Ort, C. et al. Spatial differences and temporal changes in illicit drug use in Europe quantified by wastewater analysis. Addict. Abingdon Engl. 109, 1338–1352 (2014).
Thomas, K. V. et al. Comparing illicit drug use in 19 European cities through sewage analysis. Sci. Total Environ. 432, 432–439 (2012).
Wastewater-based epidemiology and drugs topic page | www.emcdda.europa.eu. https://www.emcdda.europa.eu/topics/wastewater_en.
Prado, T. et al. Wastewater-based epidemiology as a useful tool to track SARS-CoV-2 and support public health policies at municipal level in Brazil. Water Res. 191, 116810 (2021).
WHO Coronavirus (COVID-19) Dashboard | WHO Coronavirus (COVID-19) Dashboard With Vaccination Data. https://covid19.who.int/.
Chen, J. et al. Clinical progression of patients with COVID-19 in Shanghai, China. J. Infect. 80, e1–e6 (2020).
Chen, L., Lou, J., Bai, Y. & Wang, M. COVID-19 disease with positive fecal and negative pharyngeal and sputum viral tests. Am. J. Gastroenterol. 115, 790 (2020).
Oran, D. P. & Topol, E. J. Prevalence of asymptomatic SARS-CoV-2 infection. Ann. Intern. Med. 173, 362–367 (2020).
Yang, R., Gui, X. & Xiong, Y. Comparison of clinical characteristics of patients with asymptomatic vs symptomatic coronavirus disease 2019 in Wuhan, China. JAMA Netw. Open 3, e2010182 (2020).
Yeo, C., Kaushal, S. & Yeo, D. Enteric involvement of coronaviruses: Is faecal-oral transmission of SARS-CoV-2 possible?. Lancet Gastroenterol. Hepatol. 5, 335–337 (2020).
Xiao, F. et al. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology 158, 1831-1833.e3 (2020).
Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003. https://www.who.int/publications/m/item/summary-of-probable-sars-cases-with-onset-of-illness-from-1-november-2002-to-31-july-2003.
Xu, Y. et al. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 2, 1–4. https://doi.org/10.1038/s41591-020-0817-4 (2020).
Zang, R. et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 5, 2 (2020).
Cheung, K. S. et al. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a hong kong cohort: Systematic review and meta-analysis. Gastroenterology 159, 81–95 (2020).
Wu, Y. et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 5, 434–435 (2020).
Foladori, P. et al. SARS-CoV-2 from faeces to wastewater treatment: What do we know?. A review. Sci. Total Environ. 743, 140444 (2020).
Pérez-Cataluña, A. et al. Detection Of Genomic Variants Of SARS-CoV-2 Circulating In Wastewater By High-Throughput Sequencing. https://doi.org/10.1101/2021.02.08.21251355 (2021).
Brandeburová, P. et al. Wastewater-based epidemiology to assess the occurrence of new psychoactive substances and alcohol consumption in Slovakia. Ecotoxicol. Environ. Saf. 200, 110762 (2020).
R: The R Project for Statistical Computing. https://www.r-project.org/.
Chen, Y. et al. The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J. Med. Virol. 92, 833–840 (2020).
Tim, B. et al. An alternative approach for bioanalytical assay development for wastewater-based epidemiology of SARS-CoV-2. MedRxiv https://doi.org/10.1101/2021.02.12.21251626 (2021).
La Rosa, G. et al. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 736, 139652 (2020).
Randazzo, W. et al. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 181, 115942 (2020).
Wikramaratna, P. S., Paton, R. S., Ghafari, M. & Lourenço, J. Estimating the false-negative test probability of SARS-CoV-2 by RT-PCR. Euro Surveill. Bull. Eur. Sur Mal. Transm. Eur. Commun. Dis. Bull. 25, 2 (2020).
Hata, A., Hara-Yamamura, H., Meuchi, Y., Imai, S. & Honda, R. Detection of SARS-CoV-2 in wastewater in Japan during a COVID-19 outbreak. Sci. Total Environ. 758, 143578 (2021).
Medema, G., Heijnen, L., Elsinga, G., Italiaander, R. & Brouwer, A. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environ. Sci. Technol. Lett. https://doi.org/10.1021/acs.estlett.0c00357 (2020).
Mohan, S. V., Hemalatha, M., Kopperi, H., Ranjith, I. & Kumar, A. K. SARS-CoV-2 in environmental perspective: Occurrence, persistence, surveillance, inactivation and challenges. Chem. Eng. J. 405, 126893 (2021).
Tran, H. N. et al. SARS-CoV-2 coronavirus in water and wastewater: A critical review about presence and concern. Environ. Res. 193, 110265 (2021).
Peccia, J. et al. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 38, 1164–1167 (2020).
Rooney, C. M., Moura, I. B. & Wilcox, M. H. Tracking COVID-19 via sewage. Curr. Opin. Gastroenterol. 37, 4–8 (2021).
de Aguiar-Oliveira, M. et al. Wastewater-based epidemiology (WBE) and viral detection in polluted surface water: A valuable tool for COVID-19 surveillance—A brief review. Int. J. Environ. Res. Public. Health 17, 2 (2020).
Adelodun, B., Ajibade, F. O., Ibrahim, R. G., Bakare, H. O. & Choi, K.-S. Snowballing transmission of COVID-19 (SARS-CoV-2) through wastewater: Any sustainable preventive measures to curtail the scourge in low-income countries?. Sci. Total Environ. 742, 140680 (2020).
NORMAN – SCORE joint initiative to facilitate data comparison between “SARS-CoV-2 in sewage” studies | NORMAN. http://www.normandata.eu/?q=node/361.
Lescure, F.-X. et al. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect. Dis. 20, 697–706 (2020).
Zheng, S. et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ 369, 1443 (2020).
HERA Incubator. European Commission - European Commission. https://ec.europa.eu/commission/presscorner/detail/en/fs_21_650.
Acknowledgements
We would like to thank Bratislava Water Company BVS a.s. This article was supported by the Slovak Research and Development Agency under project numbers APVV-17-0183, APVV-19-0250 and PP-COVID-20-0019. "This article was written thanks to the generous support under the Operational Program Integrated Infrastructure for the project: "Strategic research in the field of SMART monitoring, treatment and preventive protection against coronavirus (SARS-CoV-2) ", Project no. 313011ASS8, co-financed by the European Regional Development Fund." "This article was financially supported by project VIR-SCAN—Wastewater monitoring data as an early warning tool to alert COVID-19 in the population ("EOSCsecretariat.eu has received funding from the European Union's Horizon Program call H2020-INFRAEOSC- 05-2018-2019, grant Agreement number 831644.").
Author information
Authors and Affiliations
Contributions
T.M, M.T., M.Gal wrote the main manuscript text and created graphical abstract N.K., Z.T., M.Gall created the mathematical modeling, computational code and appropriate figures. A.F., J.K., A.So. performed RNA isolation and RT-qPCR analysis A.Sk., N.B., I.H., A.K., M.F. collected wastewater samples. All authors revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Krivoňáková, N., Šoltýsová, A., Tamáš, M. et al. Mathematical modeling based on RT-qPCR analysis of SARS-CoV-2 in wastewater as a tool for epidemiology. Sci Rep 11, 19456 (2021). https://doi.org/10.1038/s41598-021-98653-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-98653-x
- Springer Nature Limited
This article is cited by
-
An integrated data analysis and machine learning approach to track and monitor SARS-CoV-2 in wastewater treatment plants
International Journal of Environmental Science and Technology (2024)
-
Optimizing campus-wide COVID-19 test notifications with interpretable wastewater time-series features using machine learning models
Scientific Reports (2023)
-
20-Month monitoring of SARS-CoV-2 in wastewater of Curitiba, in Southern Brazil
Environmental Science and Pollution Research (2023)