Abstract
As one of the largest transcription factor family, basic helix-loop-helix (bHLH) transcription factor family plays an important role in plant metabolism, physiology and growth. Berry color is one of the important factors that determine grape quality. However, the bHLH transcription factor family’s function in anthocyanin synthesis of grape berry has not been studied systematically. We identified 115 bHLH transcription factors in grape genome and phylogenetic analysis indicated that bHLH family could be classified into 25 subfamilies. First, we screened six candidate genes by bioinformatics analysis and expression analysis. We found one of the candidate genes VdbHLH037 belonged to III (f) subfamily and interacted with genes related to anthocyanin synthesis through phylogenetic analysis and interaction network prediction. Therefore, we speculated that VdbHLH037 participated in the anthocyanin synthesis process. To confirm this, we transiently expressed VdbHLH037 in grape and Arabidopsis transformation. Compared with the control, transgenic materials can accumulate more anthocyanins. These results provide a good base to study the function of the VdbHLH family in anthocyanin synthesis of grape berry.
Similar content being viewed by others
Introduction
Grapes (Vitis vinifera L.) are one of the most economically valuable fruit crops in the world, mainly used in winemaking, fresh food and fruit juice1. However, the appearance of grapes is affected by uneven coloration. With the continuous improvement in people’s living standards, the requirements for grape appearance are higher. Therefore, berry color is a key factor in determining the commerciality of grape products. Anthocyanins play a vital role in the coloring process of grapes. There are more than 250 kinds of anthocyanins with known structures in plants that are classified into six main categories: cyanidin, delphinidin, pelargonidin, peonidin, petunidin and malvidin2. The grape skin is rich in anthocyanins, and the composition and content of anthocyanins generate the different berry colors. For example, petunidin and malvidin give the berry a mazarine and purple color, and the cyanidin and pelargonidin make the berry bright red. Other factors such as co-pigmentation and pH also affect the berry color3. In addition, anthocyanins play an important role in response to biotic and abiotic stresses. Castellarin et al. found that water stress induced change in gene expression regulating flavonoid biosynthesis in grape berries4. Gutha et al. found that compared with normal grape leaves, the leaves infected with leaf roll virus accumulated more anthocyanin, and the expression level of genes involved in flavonoid synthesis pathway was also significantly increased5.
Anthocyanins are important secondary metabolites belonging to the flavonoid compound that are synthesized by the condensation of anthocyanins and glycoside via glycoside bonds. They are widely found in roots, stems, flowers and fruits of plants. The anthocyanin biosynthesis pathway has more than 20 steps catalyzed by a series of enzymes. Its synthesis is regulated by structural and regulatory genes. Structural genes mainly include CHS (chalcone synthase), CHI (chalcone isomerase), F3H (flavanone3-hydroxylase), DFR (dihydroflavonol4-reductase), ANS (anthocyanidin synthase) and UFGT (flavonoid3-O-glucosyltransferase)6. These structural genes are regulated, in many plants, by MBW regulatory complex composed of R2R3 MYB TFs, bHLH protein and WD40-repeat protein7. Studies have shown that MYB is one of the most important transcription factors regulating anthocyanin biosynthesis8. The first MYB transcription factor in plants was found in maize, and was involved in the metabolic regulation of flavonoids9. MYB is the most widely distributed transcription factor family, with 138 members in grapes. It was found that MybA regulated anthocyanin synthesis through the expression of UFGT in Kyoho grape10. Kobayashi et al. found that white coloration is caused by retrotransposon insertion in MybA and that the same mutant allele has spread among most white grape cultivars in the world11. bHLH is also a major transcription factor regulating anthocyanin biosynthesis, which can directly activate DFR expression and can also promote anthocyanin accumulation by interacting with MYB. The WD40 transcription factor associated with coloration was also found in grapes. It was functionally verified and found to be expressed only on colored grapes12.
The bHLH domain consists of 50–60 amino acids that can be divided into basic region and HLH region13.The basic region at the N-terminus that contains approximately 15 amino acids is responsible for binding to DNA14. It has a highly conserved HER motif (His5-Glu9-Arg3) that recognizes a consensus hexanucleotide sequence E-box. Based on the different central bases, E-box is of different types, of which G-box is the most common type. Two amphipathic α-helices of the HLH region consisting of roughly 40 amino acids are separated by a loop of variable length15. It can promote the formation of homodimers or heterodimers through interacting with other bHLH proteins16.
The bHLH protein family is one of the largest families of TFs in plants; for example, there are 162 bHLH genes in Arabidopsis thaliana17, 192 in tobacco (Nicotiana tabacum)18, 159 in tomato (Solanum lycopersicum)19, 188 in apple (Malus × domestica)20, and 113 in strawberry (Fragaria × ananassa)21. It is usually divided into 15–26 subfamilies. For instance, 162 bHLH members of Arabidopsis are classified into 26 subfamilies. 230 bHLH proteins in Chinese cabbage (Brassica rapa ssp. pekinensis) are divided into 24 subfamilies13. Apple contains 18 subfamilies according to the phylogenetic tree analysis20. In plants, transcription factors belonging to the same subfamily have high similarity in structure, motif and protein function22. bHLH family are involved in many developmental and physiological processes in plants, such as anthocyanin biosynthesis23,24, growth and development25,26, and defense response to biotic and abiotic stress27,28. Different types of bHLH genes have different biological functions. It has been shown that bHLH III (d + e) subfamily members can enhance plant resistance and regulate anthocyanin synthesis through JA signaling pathway29,30. In addition, members of the III (f) subfamily have been proved to be involved in anthocyanin synthesis31.
The berry color of Chinese wild grapes is generally black, the spine grape has both black and white berry, which is of great significance for studying the coloring mechanism of grape berry. In this study, we analyzed 115 bHLH transcription factors comprehensively and systematically in spine grape. First, we mapped the bHLH transcription factors on to the 19 chromosomes of grape. Then we analyzed phylogenetic relationships, gene structural features, distribution of key amino acid residues in bHLH domain, conserved motifs and protein interaction network of bHLH transcription factors. Finally, six candidate genes were screened by combining with results of transcriptome sequencing and qRT-PCR validation. We verified VdbHLH037 of III (f) subfamily function by heterologous expression in Arabidopsis thaliana and transient expression in “Kyoho” grapes. The results show that VdbHLH037 plays a positive role in the regulation of anthocyanin synthesis.
Results
Identification and annotation of bHLH transcription factors in grape
A total of 137 bHLH transcription factors were obtained from EnsemblPlants database. Subsequently, we used the conserved domain to determine the existence of the conserved bHLH domain. Some members without the bHLH domain were removed. Then we analyzed the nucleotide and amino acid sequences through SMART and DNAStar. If the sequences were similar, only one member with the longest sequence was left. Finally, we screened 115 VdbHLH members for the further analysis according to the results of transcriptome sequencing15. We collated the information of 115 VdbHLH transcription factors from the Plant Transcription Factor Database (Supplementary Table S1). We mapped the VdbHLH transcription factors on the 19 chromosomes according to their distributions on the chromosome (Fig. 1). Then we renamed them from VdbHLH001 to VdbHLH108 based on their location on the chromosome 1–19 from the Plant Transcription Factor Database. The remaining six unknown chromosomal loci were renamed from VdbHLH109 to VdbHLH115 in the order of position from the minimum to maximum. We will further study the biological functions of 115 VdbHLH genes, especially for anthocyanin synthesis.
Phylogenetic analysis of the bHLH transcription factor family in grape
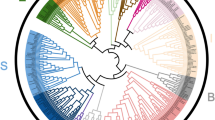
In order to better understand the function of grape bHLH protein, we need to assess their evolutionary relationship. Based on 162 AtbHLHs in Arabidopsis, we constructed an un-rooted phylogenetic tree with the nomenclature protocol according to the multiple sequence alignments of the conserved domains in grape and Arabidopsis31 (Fig. 2). On the basis of Heim’s method, we made appropriate adjustments. For example, I (a + b) subfamily was divided into I a, I b(1) and I b(2) subfamilies. III a subfamily and III c subfamily were merged into III (a + c) subfamily. Ultimately, the grape bHLH family was divided into 25 subfamilies. The members that were not assigned to the 25 subfamilies were classified as “orphans”. The plant bHLH family consists of approximately 26 subfamilies32,33, whereas there is no bHLH member in the grape X IV subfamily, probably due to the long evolutionary process. We found that the number of members of different subfamilies varies greatly. For example, the largest subfamily X II contains 14 members, while the smallest subfamilies II, VIII a, VIII c(1) and X III contain only one member. The classification of the grape bHLH family provides evidence of their evolutionary relationship.
An un-rooted phylogenetic tree of the VdbHLH family was constructed through the neighbor-joining method. The phylogenetic tree was constructed online with Interactive Tree of Life (iTOL). The numbers are bootstrap values based on 1000 iterations. Only bootstrap values larger than 50% support are displayed.
Gene structure and conserved motif analysis of VdbHLH genes
In order to obtain information about the VdbHLH family, we analyzed the structural characteristics of the VdbHLH family and labeled the phase information through GSDS2.0 (Gene Structure Display Server) (Supplementary Fig. S1). The results of the structural analysis showed that the number of exons ranged from 1 to 11. Four members had no introns. The average number of exons in 25 subfamilies ranged from 1 (VIII b) to 9 (V a). All four members without introns belong to the (VIII.
b) family. Introns can occur anywhere in the gene. If the intron is located between the third nucleotide of one codon and the first nucleotide of another codon, it is called phase 0 intron. Correspondingly, the introns between the first nucleotide and the second nucleotide of a codon are called the phase 1 introns. The introns between the second nucleotide and the third nucleotide of a codon are called the phase 2 introns. This is very important for the process of exon replication. Exons between two identical phases are called symmetric exons, whose nucleotide number is an integer multiple of three, which will not cause the reading frame to be shifted and can be copied. In contrast, asymmetric exons cannot be replicated. It is generally believed that symmetric exons and phase 0 introns can facilitate exon shuffling, recombinational fusion and protein domain exchange34,35. The statistics of 601 exons are as follows: 53 symmetrical exons between phase 0 introns, 37 symmetrical exons between phase1 intron and 52 symmetrical exons between phase 2 introns. Among the 646 introns, 190 are phase 0 introns, 133 are phase 1 introns and 323 are phase 2 introns. The results of these analyses illustrate the diversity of the VdbHLH family.
It is generally believed that motifs play an important role in the interaction of different modules in transcriptional complexes and transduction of signal15,31. In addition, the structure of a motif is closely related to gene classification. Therefore, we analyzed the 20 conservative motifs of the VdbHLH family and their distribution among different subfamilies using MEME (Supplementary Fig. S2). Furthermore, we also counted the sequence, length and number of occurrences of motifs (Supplementary Table S2). The subfamily IV a has the most types of motifs (nine types) and subfamily X III has the least types of motif (one type). The average number of motifs ranges from two (II, VIII a, VIII b, VIII c and X V) to seven (III f). All motifs occur only once per gene. Some motifs are universal, 113 members have motif2, and 96 members have motif1. Some motifs are conservative and only appear in a particular subfamily. For example: motif19 exists only in subfamily I B (2) (VdbHLH049, VdbHLH066 and VdbHLH067), motif20 exists only in subfamily III b (VdbHLH011, VdbHLH076 and VdbHLH087) and motif18 exists only in subfamily III f (VdbHLH013, VdbHLH037 and VdbHLH082). This may be why each subfamily has a specific biological function. The analysis of motif and gene structure provides a further theoretical basis for the VdbHLH subfamily classification.
The conserved amino acid residues in bHLH domain and their ability of DNA-binding
To further understand the function of the VdbHLH family, we performed multiple sequence alignment of 115 VdbHLH domains and calculated the percentage of conserved amino acids based on previous results (Fig. 3). The bHLH domain is made up of one basic region, two helix regions and one loop region. The results show that the conservation of basic region and helix region is higher than that of the loop region. The bHLH region of grape is composed of 65 amino acid residues, of which 21 conserved amino acid residues with a consensus rate of more than 50%. Among them, Arg-16, Leu-29 and Leu-65 are extremely conservative, with a consensus rate of more than 90% (Supplementary Table S3). Previous studies have shown that Glu-13, Arg-16 and Arg-17 in the basic region are critical for DNA binding, and Leu-29 and Leu-65 in the helix region are very important for dimerization activity26. By comparison, we found that seven amino acid residues (Ile-20, Leu-26, Gln-30, Met-54, Ile-59, Ile-62 and Leu-65) in plants are more conserved than those in animals, indicating that they play a very important role in plants.
The characterization and distribution of bHLH domains across all VdbHLH. (a) The sequence logo of the bHLH domain. The asterisks represent the 19 conserved amino acids mentioned previously by Atchley. The capital letters represent conserved amino acids that contain more than 50%. (b) The distribution of amino acids in the bHLH domain. The columns of different colors represent the percentage of amino acids at this site.
It is generally believed that the basic region is critical for DNA binding and the bHLH family to achieve its biological function36. According to Atchley’s research, the basic region of the bHLH protein has 13 amino acid residues, whereas the grape contains 17 amino acid residues, of which more than five are responsible for DNA binding37. The number of amino acid residues in the basic region of VdbHLH089 is less than six, which is considered incapable of DNA binding, so we designated it as a non-DNA binding protein19. As previously reported, we have divided the remaining 114 VdbHLH members into three categories: G-box (His/Lys-9, Glu-13 and Arg-17), E-box (Glu-13 and Arg-16) and non-E-box (Glu-13 and Arg-16 do not appear together)38,39. Based on the conserved amino acid residues in the basic region, we predicted 60 for the G-box-binding proteins, 27 for the non-G-box-binding proteins and 27 for the non-E-box-binding proteins. In addition, one of them was classified as a non-DNA-binding protein (Supplementary Table S4).
The collinear correlations of bHLH genes in grape, Arabidopsis and tomato
Comparative genomics is a powerful tool for quickly understanding, localizing and cloning unknown genes by comparing genes and genomic structures. The rationale is that there is interspecific and intraspecific synteny. The synteny refers to the similarity in location and sequence of genes on interspecific and intraspecific homologous chromosomes40. The synteny analysis of the genome shows a large number of collinear regions in the genome that can be used as direct evidence for the existence of whole-genome duplication (WGD)41,42. Ohno first proposed the hypothesis of polyploidization, that is, genome-wide replication, to explain the way in which diploid genomes evolve into tetraploids through WGD43. In 1997, Wolfe et al. confirmed the WGD hypothesis in Saccharomyces cerevisiae genome44. In 2000, the first plant genome-wide sketch was sequenced and analyzed. The evolutionary history of Arabidopsis genome suggests that it has undergone two whole-genome duplications (WGD: α and β) and one whole-genome triplication (WGT: γ)45. In 2007, the genome of grapes was sequenced. Studies have shown that the ancestral genome of grapes may be paleohexaploid46. The study of grape genome duplication events laid the theoretical foundation for hexaploid sharing of the ancestral genomes of dicotyledons and opened doors for the study of the evolutionary history of ancestral genomes in angiosperms. Subsequently, the whole-genome sequencing of 14 fruit trees species was completed. The whole-genome sequencing results of these fruit tree species have established a huge resource platform for fruit tree molecular biology research that not only helps in understanding their genome structure and functions but also has important guiding significance for exploring the origin and evolution of fruit trees, locating and cloning important functional genes and accelerating molecular breeding.
Comparative genomics confirmed that the grapes did not undergo any additional genome-wide replication event, while the ancestors in Solanaceae experienced an independent WGT event after differentiation47. Therefore, we carried out a few synteny analyses of grape and tomato genomes and found that the number of tomato genes was less than three times that of grapes. These results indicate that a large number of gene losses occur in WGD of tomato. We use OrthoMcl to analyze the orthologous bHLH genes in grape, Arabidopsis and tomato genome. We found 138 bHLH orthologous gene pairs between grape and tomato, but only 69 bHLH orthologous gene pairs between grape and Arabidopsis (Supplementary Table S5). The result was consistent with the close relationship between grape and tomato. In the analysis of synteny, we found each grape bHLH gene has one to three orthologous tomato genes. This shows that with the triplication of genome, some bHLH genes replicate as well. Through collinear correlations of the bHLH gene, we found 54, 10 and 75 paralogous bHLH gene pairs in grape, Arabidopsis and tomato, respectively (Supplementary Table S6). We used software Circos to show the relationships of orthologous and paralogous bHLH genes among grape, Arabidopsis and tomato (Fig. 4).
The analysis of paralogous bHLH genes and their orthologues in grape, Arabidopsis and tomato. (a) The analysis of paralogous and orthologues in grape (Chr1-Chr19) and Arabidopsis (A1-A5). The blue lines represent the paralogous bHLH genes in grape. The green lines represent the paralogous bHLH genes in Arabidopsis. The red lines represent their orthologous bHLH genes. (b) The analysis of paralogous and orthologues in grape (Chr1-Chr19) and tomato (A1-A12). The blue lines represent the paralogous bHLH genes in grape. The green lines represent the paralogous bHLH genes in tomato. The red lines represent their orthologous bHLH genes. The relationships of orthologous and paralogous bHLH genes among grape, Arabidopsis and tomato were drawn by Circos 0.69.6 (http://circos.ca/).
The expression level of spine grape berry bHLH genes at different developmental stages
We sequenced and analyzed the berry skins 40 (before veraison), 80 (at veraison) and 120 (berry ripening period) days after flowering of black and white spine grape. Among the 115 members of the bHLH family, 95 bHLH genes distributed in 21 subfamilies are expressed at least one of the developmental stages of the spine grape berry (Fig. 5). In order to screen the bHLH family candidate genes associated with anthocyanin synthesis and accumulation in pericarp of spine grape, reads per kilobase per million (RPKM) values were used to estimate the expression levels of bHLH family members. We screened 6 bHLH candidate genes (VdbHLH003, VdbHLH004, VdbHLH033, VdbHLH037, VdbHLH062 and VdbHLH097) by differential gene expression pattern analysis. These genes were mainly expressed at 80 and 120 days after the flowering of the black spine grape, and the expression level was low or not expressed in the 40 days after the flowering of the black spine grape and in the white spine grape. We validated the candidate genes by qRT-PCR, which was consistent with the results of transcriptome sequencing (Supplementary Fig. S3). Among them, the expression levels of VdbHLH033, VdbHLH037 and VdbHLH062 increased with the ripening of blackberries. According to the previous analysis, VdbHLH037 belongs to the subfamily III f. It is suggested that it may be related to anthocyanin synthesis in grapes.
The heatmap of black and white spine grape berry bHLH genes at different developmental stages. Note: B1, B2 and B3 represent black spine grape berry of 40, 80 and 120 days after anthesis, respectively. W1, W2 and W3 represent white spine grape berry of 40, 80 and 120 days after anthesis, respectively.
Interaction network prediction and functional analysis of candidate gene
Interaction network analysis helps us understand gene function and efficiency21. We used STRING to predict the interaction network of candidate genes based on VdbHLH orthologs in Arabidopsis (Fig. 6). VdbHLH003 (AMS in Arabidopsis) plays a crucial role in tapetum development and is required for male fertility and pollen differentiation, especially during the post-meiotic transcriptional regulation of microspore development within the developing anther. The interaction between VdbHLH004 (BPEp in Arabidopsis) and ARF8 has been verified in Arabidopsis. Therefore, it is believed that BPEp influences cell size and ultimately controls petal size by regulating the expression of auxin response gene. VdbHLH033 (FRU in Arabidopsis) is related to iron absorption. VdbHLH037 (TT8 in Arabidopsis) synergizes with TT1, PAP1 and TTG1 to regulate the flavonoid pathway. In addition, it also affects the expression of DFR (dihydroflavonol 4-reductase). The complex of TT8, TT2 and TTG1 is crucial to BAN genes that regulate flavonoid synthesis in seed endothelium. The genes MYB113 and MYB114 are involved in regulating anthocyanin synthesis. F3H (Flavanone 3β-hydroxylase), LDOX (Leucoanthocyanidin dioxygenase) and UF3GT (UDP glucose-flavonoid 3-O-glucosyltransferase) are enzymes that play a key role in the synthesis of anthocyanins. VdbHLH062 (ILR3 in Arabidopsis) plays a role in resistance to amide-linked indole-3-acetic acid (IAA) conjugates. The function of VdbHLH097 (AT1G22490 in Arabidopsis) is still unclear. These analyses suggest that VdbHLH037 may be involved in anthocyanin synthesis.
Overexpression of VdbHLH037 promoted anthocyanin accumulation of berry
Forty-five days after anthesis, green berry was used to inject Agrobacterium containing vector control (VC) and overexpression (OE) with a1-ml syringe. After eight days, the berries injected with OE showed faster color development than berries with VC (Fig. 7a). Compared with VC berries, OE berries had higher red color (a*), lower yellowness (b*) and similar lightness (L*) (Fig. 7c). The anthocyanin content of OE berries increased compared with VC berries (Fig. 7d). The number of red spots in OE berries was also more than that in VC berries after 8 days of infection (Supplementary Table S7). However, the number tended to be close after 16 days of injection (Supplementary Table S8). To further verify the results, RT-PCR was used to detect the infected berries. The results showed that the maker gene (hygromycin gene) could be detected 2, 4, 6, 8 days after infected by Agrobacterium containing OE and VC, but not by Agrobacterium alone (Fig. 7b). These results indicated that overexpression of VdbHLH037 could promote anthocyanin accumulation in berries.
Transient expression result of ‘Kyoho’ berries. (a) ‘Kyoho’ fruits agroinfiltrated with VC (vector control) and OE (overexpression) at 6 days after injection. (b) Hygromycin gene (resistance marker gene) of pHB vector in fruits screened by RT-PCR. (c) Color parameters (L*, a*, b*) of VC and OE berries. (d) The total anthocyanin content of VC and OE berries. RT-PCR were normalized to the expression of VlActin.
Overexpression of VdbHLH037 can promote anthocyanin accumulation and upregulation of anthocyanin synthesis related genes in transgenic Arabidopsis
The T3-generation homozygous transgenic Arabidopsis was used to further analyze the function of VdbHLH037 in anthocyanin synthesis. In four-week-old transgenic Arabidopsis, the leaves and petioles of the three OE lines (OE-2, OE-3 and OE-5) exhibited purple-black pigments when compared with VC lines (Fig. 8a). The anthocyanin content of three OE lines was also significantly higher than that of VC lines. There was no significant difference in rosette diameter and plant height between VC lines and OE lines (Fig. 8c). The transcription levels of 12 genes related to anthocyanin synthesis of VC and OE lines were analyzed. Compared with VC, the expression levels of AtCHI, AtCHS, AtF3H, AtDFR, AtLDOX (leucoanthocyanidin dioxygenase) and AtUGT78D2 in OE were significantly increased. The transcription level of OE-2 was the highest and that of OE-5 was the lowest. In VC, the expression level of these genes was low or barely detectable (Fig. 8d).
Functional analysis of VdbHLH037 in transgenic Arabidopsis. (a) The phenotype of four-week-old transgenic Arabidopsis with VC and OE. (b) The transcription level of VdbHLH037 in OE and VC transgenic Arabidopsis by qRT-PCR. (c) The total anthocyanin content of four-week-old transgenic Arabidopsis with VC and OE. (d) The transcription level of anthocyanin biosynthesis genes in transgenic Arabidopsis with VC and OE. qRT-PCR were normalized to the expression of AtUBQ3.
Discussion
As one of the most important horticultural crops in the world, grape can be used as table grape or for wine, drying, juicing and so on. Grape color is the key factor for the commerciality of berry and also a key selection index for grape breeding. The color of berry is determined by the composition and content of anthocyanin. Previous studies have shown that bHLH is involved in anthocyanin synthesis, but the function of bHLH in grape coloration is rarely reported23,24. Spine grape has strong adaptability and resistance and is the only Chinese wild grape with both black and white berry, which provides an excellent material for studying the coloring mechanism of grape.
Based on the results of ‘Pinot Noir’ sequencing, We screened 137 bHLH genes, removed the pseudogenes through the analysis of the conserved domain and finally obtained 115 bHLH genes. We named them according to their positions on chromosomes (Fig. 1). Then we conducted a phylogenetic analysis of VdbHLH and divided it into 25 subfamilies, the results are similar to Arabidopsis and tomato17,36. According to the evolutionary relationship, VdbHLH037 belongs to III (f) subfamily (Fig. 2). Members of this subfamily have been found to be related to anthocyanin synthesis. In three strawberry varieties, seven candidate bHLH genes related to anthocyanin synthesis were screened by genome-wide analysis of the bHLH family. In addition, it is considered that the FabHLH29 gene of III (f) subfamily is related to anthocyanin synthesis through gene interaction network analysis and experimental verification17. In Freesia hybrida, FhGL3L and FhTT8L of III (f) subfamily are involved in flavonoid biosynthesis48. In chrysanthemums, CmbHLH2 of III (f) subfamily regulates anthocyanin content by binding to the promoter of CmDFR and interacting with CmMYB6 so that flowers appear red, pink and yellow49. In skins and seeds of grape, VvMYC1 regulates anthocyanin and proanthocyanidin synthesis through interacting with MYB5a, MYB5b, MYBA1/A2 and MYBPA1 to adjust genes of flavonoid pathway50. Through further analysis of gene structure and conserved motif, we added evidence supporting the phylogenetic relationship of the VdbHLH family (Supplementary Figs. S1 and S2). The basic region of the VdbHLH family determines the binding ability of DNA. The ratio of E-box-binding proteins in the VdbHLH family is lower than that in Arabidopsis and tomato, which indicates that binding motifs of the VdbHLH family are diverse (Fig. 3).
Through comparative genome analysis, we found that although some bHLH genes were lost during plant evolution and genome duplication events, the number of the bHLH gene was increasing (Fig. 4). In addition, it has been reported that there are more bHLH genes in higher plants than in lower plants13. These results indicated that bHLH genes play a key role in plant evolution. In order to further understand the function of bHLH genes in grape berry coloring, we analyzed the expression pattern of the bHLH family in the three periods of black and white spine grapes. The results showed that the expression level of VdbHLH003, VdbHLH004, VdbHLH033, VdbHLH037, VdbHLH062 and VdbHLH097 was significantly increased at the veraison stage of black spine grape, and was low or almost undetectable in the early development stage of black spine grape and the three development stages of white spine grape (Fig. 5). VdbHLH037 is the highest up-regulated gene among the six genes, which was subsequently confirmed by qRT-PCR (Supplementary Fig. S3). Previously it was found that bHLH proteins perform their function by forming homodimer with bHLH protein or heterodimer with non- bHLH protein51. Therefore, we used Arabidopsis orthologs to predict the regulatory network of these six genes. The predicted interaction genes F3H, DFR, LDOX and UF3GT of the VdbHLH037 gene were involved in the synthesis of anthocyanin. Besides, the interaction between MYB transcription factor and VdbHLH037 is predicted (Fig. 6). Previous studies have shown that bHLH regulates anthocyanin biosynthesis by forming MBW transcriptional complex with MYB and WD4052. In addition, the sequence of VdbHLH037 and AtTT8 is found to be highly similar. AtTT8 belongs to III (f) subfamily and regulates anthocyanin synthesis in Arabidopsis by forming MBW transcriptional complex with AtTT2 and AtTTG153. The interaction network predicted by the remaining five genes had no direct relationship with the regulation process of anthocyanin synthesis. Accordingly, by the above results, it can be speculated that VdbHLH037 plays a positive role at the veraison period in berry.
To understand the regulatory mechanism of VdbHLH037 in anthocyanin synthesis of grape berry, we used the transient expression technology in berry to study its function54. After infection, compared with the VC, the coloration of OE berry was improved due to higher a* value. In addition, OE berries could accumulate more anthocyanin than VC. However, as time went on, the difference gradually decreased, which may be due to transient expression. Meanwhile, we also used transgenic Arabidopsis to study the function of VdbHLH037 in the synthesis of anthocyanin. The results showed that OE-2, OE-3 and OE-5 could accumulate more anthocyanin than VC. The transcription level of AtCHI, AtCHS, AtF3H, AtDFR, AtLDOX and AtUGT78D2 in OE berries was significantly higher than that in VC berries, and the expression level of these genes was consistent with the phenotype of transgenic Arabidopsis. AtCHI and AtCHS participated in the synthesis of flavonoid. AtF3H could catalyze and modify flavonoid. The flavonoid synthesis pathway entered the anthocyanin synthesis pathway under the action of AtDFR and AtLDOX. The gene AtUGT78D2 was responsible for encoding the glycosyltransferase. These results suggested that VdbHLH037 could promote anthocyanin accumulation by regulating related genes in the anthocyanin synthesis pathway. This study provides a theoretical basis for further understanding the function of bHLH family members in the process of grape berry coloring.
Methods
Identification and annotation of the grape bHLH family
The Hidden Markov Model (HMM) profile of the bHLH domain (PF00010) was downloaded from protein family (Pfam: http://pfam.sanger.ac.uk/). The PinotNoir PN40024 genome were downloaded from EnsemblPlants database (http://plants.ensembl.org/Multi/Tools/Blast?db=core) and were used as reference to blast the peptide with the BLASTp algorithm. All grape genes with non-redundant hits and expected values less than 0.001 were considered for further analysis. We obtained 137 VdbHLH genes. The 115 VdbHLH genes were screened by analyzing the conserved domain using conserved domains database (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and nucleotide and amino acid sequences using SMART (http://smart.embl-heidelberg.de/) and DNAStar. Details of 115 VdbHLH genes were obtained from the Plant Transcription Factor Database (http://planttfdb.cbi.pku.edu.cn/) and ExPASy Proteomics server (http://web.expasy.org/compute_pi/). The protein sequence of Arabidopsis and tomato was downloaded from Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html).
Bioinformatic analysis of bHLH family for grape
According to the distribution of the bHLH family on chromosome, we used MapDraw V2.1 to locate them on 19 chromosomes of grape. We analyzed the phylogenetic tree of the bHLH family by using iTOL (https://itol.embl.de/itol.cgi). The numbers are bootstrap values based on 1,000 iterations. Only bootstrap values larger than 50% support are displayed. We used GSDS2.0 (http://gsds.cbi.pku.edu.cn/) to analyze the gene structure and MEME (http://meme-suite.org/index.html) to analyze the conserved motif. Only motifs with an e-value < 1e−20 were retained for further analysis. We performed multiple sequence alignment of VdbHLH by CLUSTALW. The relationships of orthologous and paralogous bHLH genes among grape, Arabidopsis and tomato were constructed by OrthoMcl (https://orthomcl.org/orthomcl/). P-value cut-off of 1e−5 was chosen for all clusters constructed. The heatmap was drawn with Multiple experiment viewer (https://sourceforge.net/projects/mev-tm4/files/mev-tm4/). The interaction network of candidate genes was predicted by STRING (https://string-db.org/cgi/input.pl) with option value > 0.700.
Transient expression of grape and transformation of Arabidopsis
The open reading frame (ORF) of VdbHLH037 from spine grape was cloned into the BamHI site of the pHB vector with the Quick-Fusion Cloning Kit (biotool.cn) and a pair of specific primers (Supplementary Table S9). The recombinant plasmid and pHB vector were transformed into Agrobacterium tumefaciens strain GV3103. The ‘Kyoho’ were grown in China National Germplasm Resources Repository of Grape, Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences. When the culture reached an OD600 of approximately 1.0, Agrobacterium cells were harvested and resuspended in infection buffer (10 mM MgCl2, 10 mM MES, pH 5.8, and 100 µM acetosyringone) and shaken for 4 h at 28 °C before being used for infiltration. Large green grape berries (45 DAA) were used and infected with a syringe until the whole fruit became hygrophanous. Approximately 2, 4, 6, and 8 days after treatment, the fruits infiltrated with the overexpression were harvested and used to detect the vector by RT-PCR54. For analysis of phenotypes, 200 fruits were injected, with 100 fruits being transformed with VdbHLH037 and the other 100 used as the control.
The VdbHLH037 ORF was cloned between NcoI and BglII sites of the pCAMBIA3301 vector with the Quick-Fusion Cloning Kit (biotool.cn) and a pair of specific primers (Supplementary Table S9). The recombinant plasmid and pCAMBIA3301 vector were transformed into Aabidopsis Columbia ecotype by the floral dip method55. The phosphinothricin resistance and qRT-PCR were used to screen seeds (Fig. 8B). Homozygous T3-generation plants were used for further analyses.
Color measurement
The anthocyanin content in grape skins was measured after inoculation for 8 days. The leaf of four-week-old transgenic Arabidopsis with VC and OE were used to detect anthocyanin content. The samples (50 mg) was pulverized with liquid nitrogen and then each sample was homogenized in 1% (v/v) hydrochloric acid in methanol and shaken at 4 °C overnight. The anthocyanin content in grape andArabidopsis were determined by measuring the absorbance of the extract at 530 nm using an ultraviolet–visible (UV–Vis) spectrophotometer56. The color of grape berry skin was measured after inoculation for 8 days with a handheld colorimeter (CR-400, Konica Minolta, Tokyo, Japan). Color parameters were recorded as L∗(lightness), a∗(redness) and b∗(yellowness).
Analysis of qRT-PCR and RT-PCR
The ‘Kyoho’ skin of 2, 4, 6, 8 days after infected by Agrobacterium containing OE and VC were used to detect hygromycin gene. The berry skins at 40, 80 and 120 days after anthesis of black and white spine grape were used to identify candidate genes57. The RNA was extracted using TIANGEN RNAprep Pure Plant Kit (Tiangen Biotech). The four-week-old leaf of Arabidopsis was used to screen positive plants. The RNA was isolated from samples using TRIzol reagent (Invitrogen). Genomic DNA was removed by DNaseI (Thermo Scientific). The first-strand complementary DNA (cDNA) synthesis was performed with a RevertAid First-Strand cDNA Synthesis Kit (Thermo Scientific). qRT-PCR was performed in the presence of SYBR green qPCR Master Mix (Fermentas) and the amplification was performed in the Eco Real-Time PCR system (Illumina). All reactions were performed in triplicate58. The primers were designed using Oligo 7.0 and are listed in the Supplementary Table S9. The VlActin and VdActin were used as internal controls for grape. AtUBQ3 was used as internal controls for Arabidopsis.
Statistical analysis
All experiments were replicated independently at least three times, and the data were shown as mean ± SD of three independent experiments. Data were subjected to a statistical analysis according to Student’s t-test, and significant differences among the means were determined by the LSD (least significant difference), at*p < 0.05 and **p < 0.01.
References
Tu, M. et al. Expression of a grape (Vitis vinifera) bZIP transcription factor, VlbZIP36, in Arabidopsis thaliana confers tolerance of drought stress during seed germination and seedling establishment. Plant Sci. 252, 311–323 (2016).
Castellarin, S. D. et al. Colour variation in red grapevines (Vitis vinifera L.): genomic organisation, expression of flavonoid 3’-hydroxylase, flavonoid 3’, 5’-hydroxylase genes and related metabolite profiling of red cyanidin-/blue delphinidin-based anthocyanins in berry skin. BMC Genom. 7, 12 (2006).
Gauche, C., Malagoli, E. D. S. & Bordignon Luiz, M. T. Effect of pH on the copigmentation of anthocyanins from Cabernet Sauvignon grape extracts with organic acids. Scientia Agricola. 67, 41–46 (2010).
Castellarin, S. D., Matthews, M. A., Di Gaspero, G. & Gambetta, G. A. Water deficits accelerate ripening and induce changes in gene expression regulating flavonoid biosynthesis in grape berries. Planta 227, 101–112 (2007).
Gutha, L. R., Casassa, L. F., Harbertson, J. F. & Naidu, R. A. Modulation of flavonoid biosynthetic pathway genes and anthocyanins due to virus infection in grapevine (Vitis viniferaL.) leaves. BMC Plant Biol. 10, 187 (2010).
Xie, X., Zhao, J., Hao, Y. J., Fang, C. & Wang, Y. The ectopic expression of apple MYB1 and bHLH3 differentially activates anthocyanin biosynthesis in tobacco. Plant Cell, Tissue Org. Cult. PCTOC. 131, 183–194 (2017).
Pérez-Díaz, J. R. et al. New member of the R2R3-MYB transcription factors family in grapevine suppresses the anthocyanin accumulation in the flowers of transgenic tobacco. Plant Mol. Biol. 90, 63–76 (2016).
Allan, A. C., Hellens, R. P. & Laing, W. A. MYB transcription factors that colour our fruit. Trends Plant Sci. 13, 99–102 (2008).
Paz-Ares, J., Ghosal, D., Wienand, U., Peterson, P. A. & Saedler, H. The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 6, 3553–3558 (1987).
Kobayashi, S., Ishimaru, M., Hiraoka, K. & Honda, C. Myb-related genes of the Kyoho grape (Vitis labruscana) regulate anthocyanin biosynthesis. Planta 215, 924–993 (2002).
Kobayashi, S., Goto-Yamamoto, N. & Hirochika, H. Retrotransposon-induced mutations in grape skin color. Science 304, 982–982 (2004).
Ageorges, A. et al. Four specific isogenes of the anthocyanin metabolic pathway are systematically co-expressed with the red colour of grape berries. Plant Sci. 170, 372–383 (2006).
Song, X. M. et al. Genome-wide analysis of the bHLH transcription factor family in Chinese cabbage (Brassica rapa ssp. pekinensis). Mol. Genet. Genom. 289, 77–91 (2014).
Feller, A., Machemer, K., Braun, E. L. & Grotewold, E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 66, 94–116 (2011).
Toledo-Ortiz, G., Huq, E. & Quail, P. H. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15, 1749–1770 (2003).
Murre, C., McCaw, P. S. & Baltimore, D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 56, 777–783 (1989).
Pires, N. & Dolan, L. Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 27, 862–874 (2009).
Rushton, P. J. et al. Tobacco transcription factors: novel insights into transcriptional regulation in the Solanaceae. Plant Physiol. 147, 280–295 (2008).
Sun, H., Fan, H. J. & Ling, H. Q. Genome-wide identification and characterization of the bHLH gene family in tomato. BMC Genom. 16, 9 (2015).
Mao, K., Dong, Q., Li, C., Liu, C. & Ma, F. Genome wide identification and characterization of apple bHLH transcription factors and expression analysis in response to drought and salt stress. Front. Plant Sci. 8, 480 (2017).
Zhao, F. et al. Identification of basic/helix-loop-helix transcription factors reveals candidate genes involved in anthocyanin biosynthesis from the strawberry white-flesh mutant. Sci. Rep. 8, 2721 (2018).
Li, X. et al. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 141, 1167–1184 (2006).
Hou, X. J., Li, J. M., Liu, B. L. & Wei, L. Co-expression of basic helix-loop-helix protein (bHLH) and transcriptional activator-Myb genes induced anthocyanin biosynthesis in hairy root culture of Nicotiana tabacum L and Ipomea tricolor. Acta Physiol. Plant. 39, 59 (2017).
Lim, S. H., Kim, D. H., Kim, J. K., Lee, J. Y. & Ha, S. H. A radish basic helix-loop-helix transcription factor, RsTT8 acts a positive regulator for anthocyanin biosynthesis. Front. Plant Sci. 8, 1917 (2017).
Sorensen, A. M. et al. The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J. 33, 413–423 (2003).
Carretero-Paulet, L. et al. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 153, 1398–1412 (2010).
Babitha, K. C. et al. Co-expression of AtbHLH17 and AtWRKY28 confers resistance to abiotic stress in Arabidopsis. Transgenic Res. 22, 327–341 (2013).
Ji, X. et al. A bHLH gene from Tamarix hispida improves abiotic stress tolerance by enhancing osmotic potential and decreasing reactive oxygen species accumulation. Tree Physiol. 36, 193–207 (2016).
Song, S. et al. The bHLH subgroup IIId factors negatively regulate jasmonate-mediated plant defense and development. PLoS Genet. 9, e1003653 (2013).
Liu, X. J. et al. MdSnRK1. 1 interacts with MdJAZ18 to regulate sucrose-induced anthocyanin and proanthocyanidin accumulation in apple. J. Exp. Bot. 68, 2977–2990 (2017).
Heim, M. A. et al. The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20, 735–747 (2003).
Pires, N. & Dolan, L. Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 27, 862–874 (2010).
Chu, Y. et al. Genome-wide characterization and analysis of bHLH transcription factors in Panax ginseng. Acta Pharmaceut Sin. B. 8, 666–677 (2018).
Gilbert, W. The exon theory of genes. Cold Spring Harbor symposia on quantitative biology. Cold Spring Harbor Lab. Press. 52, 901–905 (1987).
Patthy, L. Intron-dependent evolution: preferred types of exons and introns. FEBS Lett. 214, 1–7 (1987).
Wang, J. et al. Genome-wide analysis of bHLH transcription factor and involvement in the infection by yellow leaf curl virus in tomato (Solanum lycopersicum). BMC Genom. 16, 39 (2015).
Atchley, W. R., Terhalle, W. & Dress, A. Positional dependence, cliques, and predictive motifs in the bHLH protein domain. J. Mol. Evol. 48, 501–516 (1999).
Ferre-D’Amare, A. R., Pognonec, P., Roeder, R. G. & Burley, S. K. Structure and function of the b/HLH/Z domain of USF. EMBO J. 13, 180–189 (1994).
Shimizu, T. et al. Crystal structure of PHO4 bHLH domain-DNA complex: flanking base recognition. EMBO J. 16, 4689–4697 (1997).
Abrouk, M. et al. Palaeogenomics of plants: synteny-based modelling of extinct ancestors. Trends Plant Sci. 15, 479–487 (2010).
Ming, R. et al. The pineapple genome and the evolution of CAM photosynthesis. Nat. Genet. 47, 1435 (2015).
Tiley, G. P., Ane, C. & Burleigh, J. G. Evaluating and characterizing ancient whole-genome duplications in plants with gene count data. Genome Biol. Evol. 8, 1023–1037 (2016).
Ohno, S. Evolution by gene duplication (Springer, Berlin, 1970).
Wolfe, K. H. & Shields, D. C. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387, 708 (1997).
Wang, X. et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43, 1035 (2011).
Jaillon, O. et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463 (2007).
Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635 (2012).
Li, Y. et al. Two IIIf clade-bHLHs from Freesia hybrida play divergent roles in flavonoid biosynthesis and trichome formation when ectopically expressed in Arabidopsis. Sci. Rep. 6, 30514 (2016).
Xiang, L. L. et al. A novel bHLH transcription factor involved in regulating anthocyanin biosynthesis in Chrysanthemums (Chrysanthemum morifolium Ramat). PloS One. 10, e0143892 (2015).
Hichri, I. et al. The basic helix-loop-helix transcription factor MYC1 is involved in the regulation of the flavonoid biosynthesis pathway in grapevine. Mol. Plant 3, 509–523 (2010).
Hernandez, J. M., Feller, A., Morohashi, K., Frame, K. & Grotewold, E. The basic helix-loop-helix domain of maize R links transcriptional regulation and histone modifications by recruitment of an EMSY-related factor. Proc. Natl. Acad. Sci. 104, 17222–17227 (2007).
Fu, Z. et al. An R3-MYB gene of phalaenopsis, MYBx1, represses anthocyanin accumulation. Plant Growth Regul. 88, 129–138 (2019).
Baudry, A., Caboche, M. & Lepiniec, L. TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J. 46, 768–779 (2006).
Gao, Z. et al. Characterization of the ABA receptor VlPYL1 that regulates anthocyanin accumulation in grape berry skin. Front. Plant Sci. 9, 592 (2018).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Li, Y. Y. et al. MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol. 160, 1011–1022 (2012).
Sun, L. et al. Transcriptome analysis of genes involved in anthocyanins biosynthesis and transport in berries of black and white spine grapes (Vitis davidii). Hereditas 153, 17 (2016).
Ni, Z., Hu, Z., Jiang, Q. & Zhang, H. GmNFYA3, a target gene of miR169, is a positive regulator of plant tolerance to drought stress. Plant Mol. Biol. 82, 113–129 (2013).
Acknowledgements
This work was supported by the scientific and technological research in Henan Province (202102110047) and (192102110153).
Author information
Authors and Affiliations
Contributions
J.Y.C. conceived the research. M.L., L.S., and R.C. performed the experiments, analyzed the data and wrote the manuscript. H.G., D.W.C., X.L.Q., and X.Z.G. provided scientific suggestions. Z.Y.W., J.F.J., and X.C.F. revised the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, M., Sun, L., Gu, H. et al. Genome-wide characterization and analysis of bHLH transcription factors related to anthocyanin biosynthesis in spine grapes (Vitis davidii). Sci Rep 11, 6863 (2021). https://doi.org/10.1038/s41598-021-85754-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-85754-w
- Springer Nature Limited
This article is cited by
-
Genome-wide identification of bHLH transcription factors and expression analysis under drought stress in Pseudoroegneria libanotica at germination
Physiology and Molecular Biology of Plants (2024)
-
Multilayered regulation of secondary metabolism in medicinal plants
Molecular Horticulture (2023)
-
Genome-wide identification and characterization of PdbHLH transcription factors related to anthocyanin biosynthesis in colored-leaf poplar (Populus deltoids)
BMC Genomics (2022)
-
Genome-wide identification and characterization of the bHLH gene family and analysis of their potential relevance to chlorophyll metabolism in Raphanus sativus L
BMC Genomics (2022)