Abstract
Tobacco bacterial wilt (TBW) caused by Ralstonia solanacearum is the most serious soil-borne disease of tobacco. However, molecular mechanism information of R. solanacearum resistance is limited to tobacco, hindering better breeding of resistant tobacco. In this study, the expression profiles of the rootstalks of Yunyan87 (susceptible cultivar) and Fandi3 (resistant cultivar) at different stages after R. solanacearum infection were compared to explore molecular mechanisms of tobacco resistance against the bacterium. Findings from gene-expression profiling indicated that the number of upregulated differentially expressed genes (DEGs) at 3 and 7 days post-inoculation (dpi) increased significantly in the resistant cultivar. WRKY6 and WRKY11 family genes in WRKY transcription factors, ERF5 and ERF15 family genes in ERFs transcription factors, and genes encoding PR5 were significantly upregulated in the resistant cultivar response to the infection. For the first time, WRKY11 and ERF15 were found to be possibly involved in disease-resistance. The Kyoto Encyclopedia of Genes and Genomes analysis demonstrated glutathione metabolism and phenylpropane pathways as primary resistance pathways to R. solanacearum infection. In the resistant cultivar, DEGs encoding CYP450, TCM, CCoAOMT, 4CL, PAL, CCR, CSE, and CADH, involved in the synthesis of plant antitoxins such as flavonoids, stilbenoids, and lignins, enriched in the phenylpropane pathway were upregulated at 3 and 7 dpi. Furthermore, a pot experiment was performed to verify the role of flavonoids in controlling TBW. This study will strongly contribute to a better understanding of molecular interactions between tobacco plants and R. solanacearum.
Similar content being viewed by others
Introduction
Ralstonia solanacearum is an important phytopathogen that attacks Solanaceae crops such as tomato, pepper, eggplant, tobacco, etc., worldwide. The disease caused by the pathogen is notoriously known as bacterial wilt with the symptom of wilting leaves or stems, vascular browning, or even death1. Tobacco bacterial wilt (TBW), caused by R. solanacearum, is the most serious soil-borne disease in tobacco plants. Most tobacco-growing countries with moist tropical or warm-temperate climates have a greater burden of this disease2. In China, the disease incidence has been gradually increasing, reaching up to 15–35%3. The increasing incidence is seriously threatening tobacco production in the 4 major tobacco-growing areas, including 14 provinces4. Thus, breeding resistant variety is considered an economical and effective measure to prevent TBW.
It is necessary to understand plant disease-resistance’s molecular mechanisms for breeding resistant variety, especially molecular breeding. Transcriptome analysis is useful to explore the molecular basis and reveal genes related to bacterial wilt resistance. For exploring the molecular mechanisms of potato-R. solanacearum interactions, 302 differentially expressed genes were identified by combining suppression subtractive hybridization and macroarray hybridization. Among them, 81 differentially expressed genes were considered R. solanacearum resistance-related genes, and these genes played putative roles in pathogen recognition, signal transduction, transcription factor functioning, hypersensitive response, systemic acquired resistance, cell rescue, and protection5. Through the transcriptome analysis of resistant and susceptible tomato cultivar after inoculation with R. solanacearum, more than 140 genes related to pathogenesis, hormone signaling, and lignin biosynthesis were increased in resistant cultivar, but no change found in susceptible cultivar6. Chen et al.7 performed global transcriptome profiling on resistant and susceptible peanut roots under R. solanacearum infection. They found that many unique genes were mainly involved in phytoalexins’ biosynthesis, particularly in the biosynthetic pathways of terpenoids and flavonoids. However, few studies have focused on transcriptomics of R. solanacearum resistance in tobacco, limiting its molecular information.
In order to resist the invasion of pathogens, plants have developed a sophisticated two-branch innate immune system. For the first branch of plant immunity, pattern-recognition receptors (PRRs) localized on the host cell surface recognize pathogen-associated molecular patterns (PAMPs) to activate PAMP-triggered immunity (PTI). However, pathogens release effectors inside the plant cell to suppress PTI. Thus the second branch of plant immunity, effector-triggered immunity (ETI) as the final defense strategy against pathogens, has been evolved in plants to recognize these effectors by nucleotide binding leucine-rich repeat (NB-LRR) resistance proteins8,9,10. In PTI, WRKY transcription factors (WRKY TFs) and ethylene-responsive TFs (ERF TFs) play important roles as regulators of plant immunity against phytopathogen, which are regulated by signaling synergistically mediated by ethylene (ET), jasmonic acid (JA), and salicylic acid (SA) signaling pathways7,11,12,13,14,15,16,17,18,19,20,21,22,23.
In this study, we used digital gene expression (DGE) profile analysis to identify and analyze gene-expression profiles of resistant and susceptible tobacco varieties infected with R. solanacearum for days 1, 3, and 7. We also screened resistance-related genes and pathway responses to R. solanacearum. This study will provide important insights into tobacco-R. solanacearum interactions, which is conducive to understand the molecular mechanisms of resistance responses to R. solanacearum in more Solanaceae crops.
Materials and methods
Tobacco materials and R. solanacearum inoculation
Tobacco cultivars Fandi3 (resistant to TBW, R) and Yunyan87 (susceptible to TBW, S) were provided by the Tobacco Research Institute of Hubei, Wuhan, China. The R. solanacearum strain HBLC5, which is highly pathogenic, was isolated from tobacco plants collected from tobacco plantations of Lichuan, Hubei, China4.
The seeds of resistant and susceptible cultivars were cultured in a greenhouse. When seedlings grew to the 5–6 leaf stage, the wounded roots of resistant and susceptible cultivars were inoculated with HBLC5 suspension (1 × 108 CFU/mL) (OD600 = 0.1), according to the method suggested by Cellier and Prior24. The inoculated tobacco seedlings were cultured in a greenhouse at 28 ± 2 °C and a humidity of 95%. Symptoms were monitored at 1, 3, 7, and 25 days post-inoculation (dpi) using the scale described by Li et al.4. The incidence (I) and disease index (DI) of TBW were calculated using the following formula: I (%) = n/N × 100%, DI = 100 × (n1 × 1 + n2 × 3 + n3 × 5 + n4 × 7 + n5 × 9)/(N × 9), where n is the number of plants with symptoms of bacterial wilt; n1, n2, n3, n4, n5 are the number of plants with symptoms 1, 3, 5, 7, and 9, respectively; and N is the total number of observations.
Sample collection
The stem base (0–1 cm above the root) of tobacco seedlings were sampled at 1, 3, and 7 dpi. The mock-inoculated tobacco seedlings of resistant and susceptible cultivars were used as controls and also sampled at the same time points. The samples were categorized into 4 groups: mock-inoculated samples of S (M-S), denoted as M-S1, M-S3, and M-S7; pathogen-inoculated samples of S (In-S), denoted as In-S1, In-S3, and In-S7; mock-inoculated samples of R (M-R), denoted as M-R1, M-R3, and M-R7; pathogen-inoculated samples of R (In-R), denoted as In-R1, In-R3, and In-R7. In total, 24 samples were collected, and the test was repeated 2 times. The stems were rinsed with deionized water, and the samples were immediately frozen in liquid nitrogen and stored at − 80 ℃ for RNA extraction.
Sample RNA extraction and detection
Total RNA from samples was extracted using the TRIzol Reagent kit (Invitrogen, ON, Canada). RNA degradation, contamination, and purity were monitored successively. Next, RNA concentration and integrity were measured. The mRNA was enriched from total RNA, PCR products were purified (AMPure XP system), and the library quality was assessed using the Agilent Bioanalyzer 2100 system.
The clustering of index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumia, San Diego, USA). Reads of 50-bp length were generated using the Illumina HiSeq 2500 sequencing platform at Novogene Bioinformatics Technology Co. Ltd., Beijing, China.
Sequencing, data processing, and reads mapping to the reference genome
Raw data (raw reads) were sequenced using Illumina HiSeq 2500. Reads containing adapter, containing ploy-N and low-quality reads from raw data were eliminated to obtain clean data (clean reads). Q30 and GC content of clean data were also calculated. Downstream analyzes were based on high-quality clean data.
The genome sequencing data of tobacco K326 on NCBI was used as a reference genome25. Clean reads were aligned to the reference genome by using TopHat v1.4.0 (http://ccb.jhu.edu/software/tophat/manual.shtml)26,27.
Screening and cluster analysis of differentially expressed genes
Genes with an adjusted p value of < 0.05 were assigned as differentially expressed genes (DEGs) by differential expression analysis (DESeq R package, 1.10.1)28. Hierarchical clustering of all DEGs was performed using the R software (v 2.15.3) (https://cran.r-project.org/index.html), displayed by Heatmap.
Gene ontology- and KEGG-enrichment analysis of DEGs
The GOseq R package was used to perform Gene Ontology (GO)-enrichment analysis of DEGs29. GO terms with a p value of < 0.05 were considered significantly enriched by DEGs. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway-enrichment analysis30 was performed to identify significantly enriched metabolic or signal transduction pathways in tobacco DEGs. Pathways with a corrected p value of < 0.05 are considered significantly enriched in DEGs as assessed by the KOBAS software (KOBAS, Surrey, UK)31.
Expression of qRT-PCR verification gene
Nine differentially regulated genes identified through DGE profiles were validated by qRT-PCR. HiScript Reverse Transcriptase (RNase H) (VAZYME, r101-01/02) was used to assist in cDNA synthesis starting from 1 μg of total RNA of resistant and susceptible cultivars at 3 and 7 dpi. Primers of selected genes were designed using the Primer Premier 5 software (PREMIER Biosoft, Palo Alto, CA, USA). Details of primer pairs are provided in Supplementary Table 1. SYBR Green Master Mix (VAZYME, Q111-02) was used for all qRT-PCR experiments in 20 μL of the reaction mix, comprising 2 × SYBR Green Master Mix (10 μL), cDNA (4 μL), PCR forward primer (100 μM; 0.4 μL), PCR reverse primer (100 μM; 0.4 μL), 50 × ROX reference dye 2b (0.4 μL), and dd H2O (4.8 μL). The reaction procedure was performed on Applied Biosystems QuantStudio. Three replicates were performed with similar results. The L25 gene was used as an internal reference for normalization32, and the relative gene-expression was calculated using the 2−△△Ct method33.
Pot experiment on controlling TBW by flavonoids
The R. solanacearum HBLC5 suspension (1 × 108 CFU/mL) (OD600 = 0.1) was inoculated into wounds at the base of tobacco (Yunyan87, 5–6 leaves) stems under greenhouse conditions. At 12-h post-inoculation (hpi), flavonoids at concentrations of 0, 1, 2, and 4 mmol/L were poured into the base of tobacco stems (10 mL per plant, 8 seedlings per treatment, 3 replicates). The occurrence of TBW was recorded at 25 dpi, and the I and DI were calculated. The control efficiency (CE) was calculated using the following formula: CE (%) = (DI—DIn)/DI0 × 100%, n = 1, 2, and 4, where DI0 is the disease index of the treatment with 0 mmol/L flavonoid solution and DIn is the disease index of the treatment with 1, 2, and 4 mmol/L of flavonoids, respectively.
Results
Disease symptoms of TBW in Yunyan87 and Fandi3
The resistance of Yunyan87 (susceptible cultivar) and Fandi3 (resistant cultivar) was determined by observing disease symptoms at 1, 3, 7, and 25 dpi and calculating I and DI (Table 1). At 3 dpi, TBW symptoms were observed in Yunyan87, with an I value of 9.09%, but not in Fandi3 (Fig. 1). At 7 dpi, I and DI values of Yunyan87 increased to 38.18% and 7.47, respectively, whereas TBW symptoms started to appear in Fandi3 (Fig. 1). At 25 dpi, I and DI values of Yunyan87 were 96.36% and 66.06, respectively, whereas the respective values increased to 34.55% and 12.73 in Fandi3. The findings indicated that Fandi3 was more resistant to TBW than Yunyan87.
The disease symptoms of the resistant (Fandi3) and susceptible (Yunyan87) cultivars induced by Ralstonia solanacearum infection. Mock-inoculated seedling (a-1), R. solanacearum-inoculated seedling (a-2), mock-inoculated opened-stem (a-3), and R. solanacearum-inoculated opened-stem (a-4) of Yunyan87 at 3 dpi. Mock-inoculated seedling (b-1), R. solanacearum-inoculated seedling (b-2), mock-inoculated opened-stem (b-3), and R. solanacearum-inoculated opened-stem (b-4) of Fandi3 at 3 dpi. Mock-inoculated seedling (c-1), R. solanacearum-inoculated seedling (c-2), mock-inoculated opened-stem (c-3), and R. solanacearum-inoculated opened-stem (c-4) of Yunyan87 at 7 dpi. Mock-inoculated seedling (d-1), R. solanacearum-inoculated seedling (d-2), mock-inoculated opened-stem (d-3), and R. solanacearum-inoculated opened-stem (d-4) of Fandi3 at 7 dpi.
Illumina HiSeq sequencing
The RNA extracted from 24 samples were sequenced using the Illumina HiSeq 2500 platform. Overall, 299,673,192 raw reads, each 50 bp in length, were generated (Supplementary Table 2). After removing adaptor sequences, duplication sequences, ambiguous reads and low-quality reads, 296,721,446 high-quality clean reads (96.81–99.82% of the raw reads) were obtained. There were 93.36–97.13% of clean reads data from 24 samples at the Q30 level of Phred-like quality scores (an error probability of 0.001). About 94.55–96.98% of total clean reads were mapped uniquely to the reference genome. The findings demonstrated that sequenced data were reliable and suitable for further DGE analysis.
Gene screening of differential expression
Venn analysis of DEGs
To identify potential genes involved in TBW resistance, all DEGs were identified in Fandi3 (R) and Yunyan87 (S) at 1, 3, and 7 dpi compared with mock-inoculated samples. The Venn diagram showed DEGs common to both cultivars or specific to either cultivar in response to inoculation (Fig. 2). At 1 dpi, 180 DEGs (p < 0.05, 129 downregulated and 51 upregulated) and 184 DEGs (p < 0.05, 77 downregulated and 107 upregulated) were identified in Fandi3 and Yunyan87, respectively (Fig. 2a, d). Of those DEGs, 3 were Fandi3-specific, 5 were common to both cultivars, 172 were DEGs (expressed but non-DEGs in Yunyan87) only in Fandi3, and 179 were DEGs (expressed but non-DEG in Fandi3) only in Yunyan87. At 3 dpi, 7450 DEGs (p < 0.05, 4588 downregulated and 2862 upregulated) and 131 DEGs (p < 0.05, 52 downregulated and 79 upregulated) were identified in Fandi3 and Yunyan87, respectively (Fig. 2b, d). Of those DEGs, 9 were Fandi3-specific, 45 were common to both cultivars, 7396 were DEGs only in Fandi3, and 86 were DEGs only in Yunyan87. At 7 dpi, 5803 DEGs (p < 0.05, 3314 downregulated and 2489 upregulated) and 275 DEGs (p < 0.05, 140 downregulated and 135 upregulated) were identified in Fandi3 and Yunyan87, respectively (Fig. 2c, d). Of those DEGs, 9 were Fandi3-specific, 121 were common to both cultivars, 5673 were DEGs only in Fandi3, and 154 were DEGs only in Yunyan87. The findings indicated that the number of DEGs in resistant cultivar (Fandi3) had considerably increased at 3 and 7 dpi, suggesting that these days may be important periods for the resistant cultivar's response to R. solanacearum.
Differentially expressed genes (DEGs) in the resistant and susceptible cultivars inducing by Ralstonia solanacearum infection. Venn diagrams of all expressed genes and DEGs in resistant and susceptible cultivars at 1 (a), 3 (b), and 7 (c) dpi. In-R versus M-R Exp and In-R versus M-R DEGs indicated, compared with mock samples (M-R), the expressed genes and DEGs in the resistant cultivar inducing by R. solanacearum infection (In-R), respectively. In-S versus M-S Exp and In-S versus M-S DEGs indicated, compared with mock samples (M-S), the expressed genes and DEGs in the susceptible cultivar inducing by R. solanacearum infection (In-S), respectively. The number of upregulated and downregulated genes among DEGs in resistant and susceptible cultivars induced by R. solanacearum infection (d). Venn diagrams were generated in R (v 2.15.3) with the VennDiagram package (v 1.6.20, https://cran.r-project.org/web/packages/VennDiagram/index.html).
Cluster analysis of DEGs
A global view of expression profiles of tobacco’s resistance responses to R. solanacearum was obtained by classifying 9831 DEGs in both cultivars at 3 and 7 dpi using hierarchical clustering analysis. The heatmap revealed that these DEGs could be divided into 4 major groups (Fig. 3): genes upregulated in Fandi3 but downregulated in Yunyan87 (group I); genes upregulated in both cultivars (group II); genes downregulated in both cultivars (group III); and genes downregulated in Fandi3 but upregulated in Yunyan87 (group IV). It was speculated that genes in groups I and II, which were upregulated in Fandi3 but downregulated in Yunyan87 (group I) or upregulated in both cultivars (group II), could be involved in resistance responses to R. solanacearum.
Expressions of differentially expressed genes in resistant and susceptible cultivars. Each column represents the log2 (In/M) at indicated times, where In and M are read count values of the gene in inoculated and mock samples, respectively. Red and blue represent upregulated and downregulated genes, respectively. The darker the color, the higher is the degree of upregulation or downregulation. Heatmap was generated in R (v 2.15.3) with the ComplexHeatmap package (v 2.11, https://bioconductor.org/packages/release/bioc/html/ComplexHeatmap.html).
Analysis of DEGs by GO-enrichment
To investigate biological processes and functions of DEGs possibly involved in tobacco’s resistance responses to R. solanacearum, we performed GO-enrichment analysis (molecular functions, biological processes, and cell components) to classify the functions of DEGs in groups I and II. At 3 dpi, 17 GO terms were significantly enriched (p ≤ 0.05), in which 2, 6, and 9 GO terms were classified as molecular functions, biological processes, and cell components, respectively (Fig. 4a). Of those, GO terms “intracellular organelle” and “regulation of metabolic process” were involved in most DEGs. At 7 dpi, 16 GO terms were significantly enriched (p ≤ 0.05), in which 2, 10, and 4 GO terms were classified as molecular functions, biological processes, and cell components, respectively (Fig. 4b). Of those, GO terms “organic substance metabolic process,” “primary metabolic process,” and “cellular metabolic process” were involved in more than 800 DEGs, respectively.
KEGG annotation and pathway-enrichment analysis of DEGs
KEGG pathway analysis of DEGs
To further investigate the biosynthetic and signaling pathways of DEGs that could be involved in tobacco’s resistance responses to R. solanacearum, we performed pathway-enrichment analysis using KEGG. A total of 6 and 10 KEGG pathways (p ≤ 0.05) were involved in groups I and II DEGs (Table 2). These pathways were primarily involved in the phenylpropane pathway (phenylpropanoid biosynthesis, sly00940; flavonoid biosynthesis, sly00941; stilbenoid, diarylheptanoid, and gingerol biosynthesis, sly00945) and glutathione metabolism (sly00480)34,35,36.
Differential expression analysis of genes related to the KEGG pathway
Phenylalanine is a precursor of several secondary metabolites in plants. In the phenylpropanoid biosynthesis pathway, phenylalanine is transformed into p-coumaroyl CoA by phenylalanine ammonia-lyase (PAL) and 4-coumaric acid CoA ligase (4CL). Finally, through a series of biochemical reactions, p-coumaroyl CoA is converted to flavonoids, stilbenoids, and lignins (Supplementary Figs. 1–3). Our results demonstrated that 38 upregulated DEGs (0.79–5.38 folds) in Fandi3 were enriched in the phenylpropane pathway at 3 dpi, in which the expression levels of 23 genes (23/38, 60.53%) were higher in Fandi3 than in Yunyan87. At 7 dpi, 46 upregulated DEGs (1.49–8.92 folds) in Fandi3 were enriched in the phenylpropane pathway, in which the expression levels of 38 genes (38/46, 82.61%) were higher in Fandi3 than in Yunyan87 (Fig. 5, Supplementary Table 3). Moreover, 20 upregulated DEGs in Fandi3 involved in the phenylpropane pathway were identified at 3 and 7 dpi. They were related to PAL, 4CL, trans-cinnamate 4-monooxygenase (TCM), cytochrome P450 (CYP450), caffeoyl-CoA O-methyltransferase (CCoAOMT), cinnamoyl-CoA reductase (CCR), caffeoylshikimate esterase (CSE), and cinnamyl alcohol dehydrogenase (CADH). The upregulated DEGs may increase the accumulation of flavonoids, stilbenoids, and lignins, enhancing the plant’s resistance against R. solanacearum.
Comparison of the expression of genes associated with the phenylpropane pathway in resistant and susceptible cultivars in response to Ralstonia solanacearum infection. The bar represents log2 (In-R/In-S), where In-R and In-S are read count values of the gene in resistant and susceptible cultivars induced by R. solanacearum. Comparison of the expression of genes in resistant and susceptible cultivars in response to R. solanacearum infection at 3 dpi (a) and 7 dpi (b). Δ indicated common differentially expressed genes enriched in the flavonoid biosynthesis and stilbenoid, diarylheptanoid, and gingerol biosynthesis pathway at 3 and 7 dpi. Filled triangle indicated common differentially expressed genes enriched in the phenylpropanoid biosynthesis pathway at 3 and 7 dpi.
In the glutathione metabolic pathway (Supplementary Fig. 4), 22 (0.77–6.02 folds) and 38 (1.76–10.50 folds) upregulated DEGs in Fandi3 were enriched at 3 and 7 dpi, respectively (Fig. 6, Supplementary Table 4). Moreover, 6 upregulated DEGs involved in the glutathione metabolic pathway were identified at both 3 and 7 dpi, and they were associated with L-ascorbate peroxidase (APX), glutathione S-transferase (GST), and spermidine synthase (SPDE).
Comparison of the expression of genes associated with glutathione metabolism in resistant and susceptible cultivars in response to Ralstonia solanacearum infection. The bar represents log2 (In-R/In-S), where In-R and In-S are read count values of the gene in resistant and susceptible cultivars induced by R. solanacearum. Comparison of the expression of genes in resistant and susceptible cultivars in response to R. solanacearum infection at 3 dpi (a) and 7 dpi (b). Δ indicated common differentially expressed genes enriched in the glutathione metabolism at 3 and 7 dpi.
Transcription factor and PR-related gene-expression
WRKY TFs, ERFs, and pathogenesis-related proteins (PRs) play key roles in plant resistance against pathogens, implying the importance of investigating them at 3 and 7 dpi in this study.
WRKY TFs are the largest family of proteins in plants that regulate a series of defense processes and play important roles in modulating the transcription of resistance-related genes. In this study, the expressions of 55 WRKY genes were identified in both cultivars (Fig. 7, Supplementary Table 5). Of these, 15 WRKY genes (WRKY6, WRKY11, WRKY23, WRKY28, WRKY33, WRKY41, WRKY49, and WRKY65 family genes) were significantly upregulated in Fandi3 at 3 dpi, and 18 WRKY genes (WRKY6, WRKY11, WRKY15, WRKY21, WRKY40, WRKY41, WRKY50, WRKY51, WRKY70, and WRKY75 family genes) were significantly upregulated in Fandi3 at 7 dpi. Notably, 3 genes, gene_14657, gene_31173, and gene_6736 (WRKY11 family genes), were significantly upregulated in Fandi3 at both 3 and 7 dpi, and their expression levels were higher in Fandi3 than in Yunyan87. It is speculated that genes involved in WRKY11 may play an important role in tobacco resistance against R. solanacearum. Moreover, the expression levels of genes in the WRKY6 family (except gene_41493) could be important positive regulators of resistance against R. solanacearum in pepper13 identified only at 3 dpi to be higher in Fandi3 than in Yunyan87. This indicated that WRKY6 family genes might respond to R. solanacearum infection, especially in the early stage.
Comparison of the expression of genes associated with WRKY transcription factors in resistant and susceptible cultivars in response to Ralstonia solanacearum infection. The bar represents log2 (In-R/In-S), where In-R and In-S are read count values of the gene in resistant and susceptible cultivars induced by R. solanacearum.
ERFs have important biological functions in various life activities such as plant growth, development, and response to the environment. In this study, the expression of 13 ERFs genes was identified in both cultivars (Fig. 8, Supplementary Table 5). Of those, in Fandi3, 5 ERF genes (ERF3, ERF5, ERF10, and ERF15 family genes) were significantly upregulated at 3 dpi, and 5 (ERF10, ERF15, and ERF71 family genes) were significantly upregulated at 7 dpi. Moreover, gene_14860 and gene_19445 (ERF15 family genes) were significantly upregulated in Fandi3 at both 3 and 7 dpi, and their expression levels were higher than that in Yunyan87. These findings suggested that these 2 genes could be important positive regulators of tobacco resistance against R. solanacearum. Further, the expression level of gene_11665 in the ERF5 family, which could upregulate the expression levels of a series of defense-related genes in plants22, was identified only at 3 dpi and was higher in Fandi3 than in Yunyan87. This result implied that this gene might also respond to R. solanacearum infection primarily in the early stage.
Comparison of the expression of genes associated with ERF transcription factors and pathogenesis-related proteins (PR) in resistant and susceptible cultivars in response to Ralstonia solanacearum infection. The bar represents log2 (In-R/In-S), where In-R and In-S are read count values of the genes in resistant and susceptible cultivars induced by R. solanacearum.
PRs are induced by phytopathogens and defense-related signaling molecules. They often serve as diagnostic molecular markers of defense signaling pathways. PRs are effective against multiple biotic agents such as fungi, bacteria, or even insects37,38. Overall, 8 PR genes were identified in both cultivars (Fig. 8, Supplementary Table 5). In Fandi3, one PR gene was significantly upregulated at 3 dpi, and 4 PR genes were significantly upregulated at 7 dpi. Of those, genes encoding PR5 were significantly upregulated in Fandi3, and the expression levels of the genes were higher than that in Yunyan87. The findings suggested that PR5 could be a key positive regulator in tobacco’s defense against R. solanacearum infection.
Expression verification of genes associated with resistance against TBW
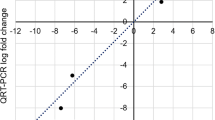
Nine DEGs that could be essential for resistance were selected to determine the relative expression level by qRT-PCR at 3 and 7 dpi. The genes were one gene (gene_34916) associated with CYP450, one gene (gene_23271) associated with CCoAOMT, 2 genes (gene_2681 and gene_64101) associated with PAL, one gene (gene_56617) associated with APX, 2 genes (gene_41493 and gene_31173) associated with WRKY TFs, and 2 genes (gene_11665 and gene_19445) associated with ERFs. The results revealed that all 9 genes were upregulated at 3 and 7 dpi in Fandi3 but had lower expression levels or downregulated in Yunyan87 (Fig. 9a). The correlation coefficient between DGE and qRT-PCR data was significant at 0.835 (p < 0.01), indicating that qRT-PCR data were consistent with DGE data and confirmed expression patterns of these genes as revealed by DGE data (Fig. 9b).
Verification of the digital gene expression (DGE) profile. Expression levels of the candidate differentially expressed genes obtained by qRT-PCR, calculated using the 2−△△Ct method with normalization to the internal reference L25 gene (a). The linear correlation between DGE profiles and qRT-PCR data (b). A correlation coefficient of 0.835 indicates an excellent linear correlation between the DGE profile and qRT-PCR data.
Control efficiency of flavonoids on TBW
KEGG-enrichment analysis of DEGs demonstrated that genes related to plant protectin synthesis in the phenylpropane pathway were significantly upregulated in Fandi3. To verify whether flavonoids were involved in resistance against TBW, flavonoids at various concentrations were applied on the stem base of Yunyan87 inoculated with 1 × 108 CFU/mL of R. solanacearum under greenhouse conditions. The DI of tobacco seedlings treated with 1, 2, and 4 mmol/L flavonoids were significantly lower than those treated with 0 mmol/L flavonoids at 25 dpi. The control efficiency values of flavonoids at 1, 2, and 4 mmol/L on bacterial wilt were 56.10%, 59.76%, and 84.15%, respectively (Table 3).
Discussion
To study the mechanism of the tobacco plant’s resistance against TBW, DEGs at different times were analyzed after inoculation with R. solanacearum. At 1 dpi, the number of DEGs in both cultivars was fewer, indicating that the resistance response to TBW was not activated. However, resistant-related genes were quickly activated in Fandi3, with the number of upregulated genes being 36.23 and 18.44 folds higher in Fandi3 than in Yunyan87 at 3 and 7 dpi, respectively. This finding is consistent with previous studies7,39 and demonstrated that Fandi3 (resistant cultivar) had a significant resistance response to TBW at 3 and 7 dpi.
Certain metabolic pathways play essential roles in plant resistance against TBW. Glutathione (GSH) is the most abundant antioxidant in cells and is crucial for life processes by protecting DNA, proteins, and other biomolecules against oxidative damage and heavy metal ions, which is favorable to the resistance against environmental stress40,41,42. Avinash et al.43 demonstrated that the expression of resistance genes associated with the ascorbic acid (AsA)-GSH pathway increased by 2.5–3.5 folds in the resistant eggplant cultivars after infection with R. solanacearum. In our study, after infection with R. solanacearum, the expression of genes associated with the GSH metabolism pathway increased significantly in Fandi3. The genes associated with APX increased by 2.16 and 3.61 folds at 3 and 7 dpi, respectively, and genes associated with GST increased by 4.15 and 4.01 folds at 3 and 7 dpi, respectively. APX is the first step of the AsA-GSH cycle and is an effective antioxidant enzyme that assists in removing H2O2 from cells44. GST is an important antioxidant enzyme related to GSH metabolism and controls the binding of GSH to certain biomolecules45. Therefore, the upregulated genes associated with APX and GST in plants could enhance the redox ability and protect cell membranes from damage caused by reactive oxygen species (ROS).
The phenylpropane pathway plays an essential role in plant disease-resistance. In the present study, genes associated with PAL and 4CL were significantly upregulated in Fandi3, which is in line with the study by Ishihara et al.6. They demonstrated that genes associated with 4CL were significantly upregulated in resistant tomato cultivars after infection with R. solanacearum, indicating that the infection could lead to the accumulation of phenylpropane derivatives in resistant cultivars. There are 2 main branching pathways in the phenylpropanoid biosynthesis pathway: the lignin pathway and the phenols and flavonoids pathway. Lignin is an important component of the plant secondary cell wall. Certain enzymes such as CADH, CSE, and CCoAOMT play central roles in lignin synthesis46,47. In the present study, genes associated with CADH, CSE, and CCoAOMT were significantly upregulated in Fandi3 after infection, consistent with the response of resistant tomato cultivars after infection with R. solanacearum6. This could be because bacterial wilt could induce lignin accumulation in plants as a physical barrier for resistance against the infection. Flavonoids and astragalus compounds as protective agents can inhibit several pathogenic bacteria48. In this study, the expression of genes associated with the biosynthesis of flavonoids, stilbenes, diarylheptanoids, and gingerol increased significantly in Fandi3, indicating that the infection of bacterial wilt in resistant variety could promote the accumulation of flavonoids and stilbenes to inhibit the reproduction of R. solanacearum and improve resistance against the infection. This finding is consistent with the conclusion of Chen et al. that bacterial wilt disease promoted the biosynthesis of flavonoids and terpenoids in resistant peanut lines49. CYP450 is an essential metabolic enzyme present in most organisms. It is mainly involved in metabolic pathways of hormones, phenylpropane, alkaloids, and terpenes50,51,52,53. In this study, genes associated with CYP450 in the phenylpropane pathway were significantly upregulated in Fandi3, indicating that CYP450 could be mainly involved in the accumulation of phenylpropane derivatives to improve the resistance of tobacco to bacterial wilt.
Yang et al. demonstrated that rice, tobacco, and Arabidopsis thaliana pretreated with flavonoids had enhanced resistance to Xanthomonas oryzae pv. oryzae, R. solanacearum, and Pseudomonas syringae pv. tomato, respectively54. Moreover, certain PR-related genes such as PR1a, NOA1, and rbohB were significantly upregulated in tobacco leaves treated with flavonoids. In this study, KEGG-enrichment analysis of DEGs revealed that genes related to plant protectin synthesis in the phenylpropane pathway were significantly upregulated in Fandi3, indicating the possible involvement of flavonoids in resistance against TBW. We, therefore, performed a pot experiment to evaluate the role of flavonoids in controlling TWB. The control efficiency of 1–4 mmol/L flavonoids on controlling TBW was 56.10–84.15%. As such, the reasons for the exogenous application of flavonoids improving resistance against TBW could be flavonoid-induced upregulation of PR genes, but this aspect requires further evidence.
TFs are the most important regulatory genes in plants. WRKY TFs play essential roles in the immune response of plants to various biological stresses55. Many studies have shown that WRKY40, WRKY6, WRKY27, and WRKY22 have positive regulatory effects on the resistance of Solanaceae crops to bacterial wilt14,15,16,17,18], and CaWRKY6 regulated CaWRKY40 to coordinate the response to bacterial wilt15. In this study, WRKY6 had a resistance response to TBW, mainly in the early stage (3 dpi), which is consistent with the response pattern of pepper to bacterial wilt15. Moreover, WRKY11 family genes demonstrated apparent positive regulation in Fandi3 and negative regulation in Yunyan87, suggesting these genes could also be important positive regulators of tobacco resistance against R. solanacearum56,57, which needs further verification. ERFs are a unique class of TFs in plants involved in response to biological and abiotic stresses58,59. Zhang et al.60 reported that the expression of ERF15 could be induced by P. syringae pv. tomato and Botrytis cinerea infection and ERF15 overexpression significantly increased resistance in plants. In this study, the expression of ERF15 family genes was significantly upregulated (3.74 and 3.58 folds at 3 and 7 dpi, respectively) in Fandi3 after infection, indicating that the gene is an important positive regulator of tobacco resistance against R. solanacearum. The expression pattern of ERF5 (gene_11665) in Fandi3 was distinct from that in Yunyan87, especially in the early stage (3 dpi). It could have regulated a series of defense genes to enhance the resistance of tobacco22. PRs are generally considered important for the systemic acquisition of disease-resistance (SAR)61. Dahal et al.62 pointed out that PR5 had a positive regulatory effect on tomato resistance against bacterial wilt. In this study, PR5-related genes demonstrated significant regulatory differences between Fandi3 and Yunyan87 after infection, in which the genes in Fandi3 were upregulated by 1.31 and 2.38 folds on average at 3 and 7 dpi after infection, respectively, whereas genes in Yunyan87 were downregulated. The results indicated the possible involvement of PR5 in tobacco resistance against R. solanacearum.
The 9 genes in Fandi3 and Yunyan87 were further analyzed using qRT-PCR. The genes were either upregulated in Fandi3 at both 3 and 7 dpi or demonstrated a distinct expression pattern in Fandi3 and Yunyan87 in DGE. Further, these genes involved in key pathways (phenylpropane pathway and glutathione metabolism) were related to PAL, CYP450, CCoAOMT, and APX or were WRKY genes and ERFs genes. Therefore, it was necessary to select these genes for verifying the DEGs identified and confirmed the expression patterns of these genes in Fandi3 and Yunyan87 by qRT-PCR.
Conclusions
In this study, the DGE sequencing technology was used to analyze DEGs and related pathways in resistant and susceptible tobacco cultivars. Both cultivars had similar transcriptome levels at the early stage of infection, but resistance-related DEGs had distinct genotype-specific expression patterns in response to TBW after 3 dpi. Meanwhile, glutathione metabolism and phenylpropane pathway were the main resistant pathways of tobacco’s response to R. solanacearum. Furthermore, a pot experiment suggested that flavonoids help control TWB. The study results reveal the molecular mechanisms involved in tobacco resistance to R. solanacearum and serve as an important guide for selecting and breeding tobacco plants resistant to TBW.
References
Grimault, V., Anais, G. & Prior, P. Distribution of Pseudomonas solanacearum in the stem tissues of tomato plants with different levels of resistance to bacterial wilt. Plant Pathol. 43, 663–668 (1994).
Denny, T. Plant pathogenic Ralstonia species. In Plant-associated bacteria (ed. Gnanamanickam, S. S.) 573–644 (Springer, Dordrecht, 2006).
Jiang, G. et al. Bacterial wilt in China: History, current status, and future perspectives. Front. Plant Sci. 8, 1549–1558. https://doi.org/10.3389/fpls.2017.01549 (2017) (PMID: 28955350).
Li, Y. et al. Genetic diversity and pathogenicity of Ralstonia solanacearum causing tobacco bacterial wilt in China. Plant Dis. 100, 1288–1296. https://doi.org/10.1094/PDIS-04-15-0384-RE (2016).
Li, G. et al. Gene transcription analysis during interaction between potato and Ralstonia solanacearum. Russ. J. Plant Physiol. 57, 685–695. https://doi.org/10.1134/s1021443710050122 (2010).
Ishihara, T., Mitsuhara, I., Takahashi, H. & Nakaho, K. Transcriptome analysis of quantitative resistance-specific response upon Ralstonia solanacearum infection in tomato. PLoS ONE 7, e46763. https://doi.org/10.1371/journal.pone.0046763 (2012).
Chen, Y. et al. Dynamics in the resistant and susceptible peanut (Arachis hypogaea L.) root transcriptome on infection with the Ralstonia solanacearum. BMC Genom. 15, 1078. https://doi.org/10.1186/1471-2164-15-1078 (2014).
Jones, J. D. & Dangl, J. L. The plant immune system. Nature 444(7117), 323–329 (2006).
Boller, T. & Felix, G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406 (2009).
Wang, G. P. et al. Resistance against Ralstonia solanacearum in tomato depends on methionine cycle and γ-aminobutyric acid metabolic pathway. Plant J. 97, 1032–1047. https://doi.org/10.1111/tpj.14175 (2019).
Huang, P. Y. et al. NINJA-associated ERF19 negatively regulates Arabidopsis pattern-triggered immunity. J Exp Bot https://doi.org/10.1101/180059 (2017).
Wang, J. H. et al. Apple ethylene response factor MdERF11 confers resistance to fungal pathogen Botryosphaeria dothidea. Plant Sci. https://doi.org/10.1016/j.plantsci.2019.110351 (2019).
McGrath, K. C. et al. Repressor-and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol. 139, 949–959 (2005).
Ashraf, M. F. et al. Capsicum annuum HsfB2a positively regulates the response to Ralstonia solanacearum infection or high temperature and high humidity forming transcriptional cascade with CaWRKY6 and CaWRKY40. Plant Cell Physiol. 59, 2608–2623. https://doi.org/10.1093/pcp/pcy181 (2018) (PMID: 30169791).
Cai, H. et al. CaWRKY6 transcriptionally activates CaWRKY40, regulates Ralstonia solanacearum resistance, and confers high-temperature and high-humidity tolerance in pepper. J. Exp. Bot. 66, 3163–3174. https://doi.org/10.1093/jxb/erv125 (2015).
Dang, F. et al. Overexpression of CaWRKY27, a subgroup IIe WRKY transcription factor of Capsicum annuum, positively regulates tobacco resistance to Ralstonia solanacearum infection. Physiol. Plant. 150, 397–411. https://doi.org/10.1111/ppl.12093 (2014).
Dang, F. et al. CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ. 36, 757–774. https://doi.org/10.1111/pce.12011 (2013).
Hussain, A. et al. CaWRKY22 acts as a positive regulator in pepper response to Ralstonia Solanacearum by constituting networks with CaWRKY6, CaWRKY27, CaWRKY40 and CaWRKY58. Int. J. Mol. Sci. 19, 1426–1444. https://doi.org/10.3390/ijms19051426 (2018) (PMID: 29747470).
Wang, Y. et al. CaWRKY58, encoding a group I WRKY transcription factor of Capsicum annuum, negatively regulates resistance to Ralstonia solanacearum infection. Mol. Plant Pathol. 14, 131–144. https://doi.org/10.1111/j.1364-3703.2012.00836.x (2013).
Berrocal-Lobo, M., Molina, A. & Solano, R. Constitutive expression of ETHYLENE RESPONSE FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 29, 23–32 (2002).
Zhang, H. B. et al. Tomato stress-responsive factor TSRF1 interacts with ethylene responsive element GCC box and regulates pathogen resistance to Ralstonia solanacearum. Plant Mol. Biol. 55, 825–834 (2004).
Lai, Y. et al. Overexpression of a pepper CaERF5 gene in tobacco plants enhances resistance to Ralstonia solanacearum infection. Funct. Plant Biol. 41, 758–767. https://doi.org/10.1071/FP13305 (2014).
Lai, Y. et al. Overexpression of a Chinese cabbage BrERF11 transcription factor enhances disease resistance to Ralstonia solanacearum in tobacco. Plant Physiol. Biochem. 62, 70–78. https://doi.org/10.1016/j.plaphy.2012.10.010 (2013).
Cellier, G. & Prior, P. Deciphering phenotypic diversity of Ralstonia solanacearum strains pathogenic to potato. Phytopathology 100, 1250–1261. https://doi.org/10.1094/PHYTO-02-10-0059 (2010).
Sierro, N. et al. The tobacco genome sequence and its comparison with those of tomato and potato. Nat. Commun. 5, 3833–3841. https://doi.org/10.1038/ncomms4833 (2014).
Trapnell, C., Pachter, L. & Salzberg, S. L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25(9), 1105–1111. https://doi.org/10.1093/bioinformatics/btp120 (2009).
Kim, D. et al. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36. https://doi.org/10.1186/gb-2013-14-4-r36 (2013).
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106. https://doi.org/10.1186/gb-2010-11-10-r106 (2010).
Young, M. D., Wakefield, M. J., Smyth, G. K. & Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 11, R14. https://doi.org/10.1186/gb-2010-11-2-r14 (2010).
Kanehisa, M. et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–D484 (2008).
Mao, X., Cai, T., Olyarchuk, J. G. & Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a vontrolled vocabulary. Bioinformatics 21(19), 3787–3793 (2005).
Schmidt, G. W. & Delaney, S. K. Stable internal reference genes for normalization of real-time RT-PCR in tobacco (Nicotiana tabacum) during development and abiotic stress. Mol. Genet. Genom. 283, 233–241. https://doi.org/10.1007/s00438-010-0511-1 (2010).
Livak, K. & Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2-△△Ct Method. Methods 408, 402–408. https://doi.org/10.1006/meth.2001.1262 (2001).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951 (2019).
Kanehisa, M., Furumichi, M., Sato, Y., Ishiguro-Watanabe, M. & Tanabe, M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. https://doi.org/10.1093/nar/gkaa970 (2021).
Kushalappa, A., Yogendra, K. & Karre, S. Plant innate immune response: Qualitative and quantitative resistance. Crit. Rev. Plant Sci. 35, 38–55 (2016).
Sajad, A. et al. Pathogenesis-related proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiol. Res. https://doi.org/10.1016/j.micres.2018.04.008 (2018).
Ghareeb, H. et al. Transcriptome of silicon-induced resistance against Ralstonia solanacearum in the silicon non-accumulator tomato implicates priming effect. Physiol. Mol. Plant Pathol. 75, 83–89. https://doi.org/10.1016/j.pmpp.2010.11.004 (2011).
Cobbett, C. & Goldsbrough, P. Phytochelatins and metallothioneins: Roles in heavy metal detoxifcation and homeostasis. Annu. Rev. Plant Biol. 53, 159–182. https://doi.org/10.1146/annurev.arplant.53.100301.135154 (2002).
Freeman, J. L. et al. Increased glutathione biosynthesis plays a role in nickel tolerance in thlaspi nickel hyperaccumulators. Plant Cell 16, 2176–2191. https://doi.org/10.1105/tpc.104.023036 (2004) (PMID: 15269333).
Mendozacózatl, D., Lozatavera, H., Hernandeznavarro, A. & Morenosanchez, R. Sulfur assimilation and glutathione metabolism under cadmium stress in yeast, protists and plants. FEMS Microbiol. Rev. 29, 653–671. https://doi.org/10.1016/j.femsre.2004.09.004 (2005).
Avinash, P., Umesha, S. & Marahel, S. Role of hydrogen peroxide and ascorbate-glutathione pathway in host resistance to bacterial wilt of eggplant. Russ. J. Plant Physiol. 64, 375–385. https://doi.org/10.1134/S1021443717030049 (2017).
Drążkiewicz, M., Skórzyńska-Polit, E. & Krupa, Z. Response of the ascorbate-glutathione cycle to excess copper in Arabidopsis thaliana (L.). Plant Sci. 164, 195–202. https://doi.org/10.1016/S0168-9452(02)00383-7 (2003).
Dixon, D. P., Fordham-Skelton, A. P. & Edwards, R. Redox regulation of a soybean tyrosine-specific protein phosphatase. Biochemistry 44, 7696–7703. https://doi.org/10.1021/bi047324a (2005) (PMID: 15909984).
Ye, Z. Association of caffeoyl coenzyme a 3-o-methyltransferase expression with lignifying tissues in several dicot plants. Plant Physiol. 115, 1341–1350. https://doi.org/10.1104/pp.115.4.1341 (1997).
Vanholme, R., Demedts, B., Morreel, K., Ralph, J. & Boerjan, W. Lignin biosynthesis and structure. Plant Physiol. 153, 895–905. https://doi.org/10.1104/pp.110.155119 (2010).
Ahuja, I., Kissen, R. & Bones, A. Phytoalexins in defense against pathogens. Trends Plant Sci. 17, 1360–1385. https://doi.org/10.1016/j.tplants.2011.11.002 (2012).
Chen, Y. et al. Alteration of gene expression profile in the roots of wild diploid Arachis duranensis inoculated with Ralstonia solanacearum. Plant. Pathol. 63, 803–811. https://doi.org/10.1111/ppa.12158 (2014).
Boachon, B. et al. CYP76C1 (Cytochrome P450)-mediated linalool metabolism and the formation of volatile and soluble linalool oxides in Arabidopsis flowers: A strategy for defense against floral antagonists. Plant Cell 27, 2972–2990. https://doi.org/10.1105/tpc.15.00399 (2015).
Han, J., Hwang, H., Choi, S., Kim, H. & Choi, Y. Cytochrome P450 CYP716A53v2 catalyzes the formation of protopanaxatriol from protopanaxadiol during ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 53, 1535–1545. https://doi.org/10.1093/pcp/pcs106 (2012).
Han, J., Kim, H., Kwon, Y. & Choi, Y. The Cyt P450 enzyme CYP716A47 catalyzes the formation of protopanaxadiol from dammarenediol-II during ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 52, 2062–2073. https://doi.org/10.1093/pcp/pcr150 (2011).
Li, H. et al. Cytochrome P450 family member CYP704B2 catalyzes the ω-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. Plant Cell 22, 173–190. https://doi.org/10.1105/tpc.109.070326 (2010).
Yang, W. et al. Rutin-mediated priming of plant resistance to three bacterial pathogens initiating the early SA signal pathway. PLoS ONE 11, e0146910. https://doi.org/10.1371/journal.pone.0146910 (2016) (PMID: 26751786).
Chen, F. et al. The WRKY transcription factor family in model plants and crops. Crit. Rev. Plant Sci. 36, 311–335. https://doi.org/10.1080/07352689.2018.1441103 (2018).
Ali, M. A. et al. Transcription factors WRKY11 and WRKY17 are involved in abiotic stress responses in Arabidopsis. J. Plant Physiol. 226, 12–21. https://doi.org/10.1016/j.jplph.2018.04.007 (2018).
Ali, M. A., Wieczorek, K., Kreil, D. P. & Bohlmann, H. The beet cyst nematode Heterodera schachtii modulates the expression of WRKY transcription factors in syncytia to favour its development in Arabidopsis roots. PLoS ONE 9, e102360. https://doi.org/10.1371/journal.pone.0102360 (2014).
Lata, C. et al. Genome-wide investigation and expression profiling of AP2/ERF transcription factor superfamily in foxtail millet (Setaria italica L.). PLoS ONE 9, e113092. https://doi.org/10.1371/journal.pone.0113092 (2014).
Li, M. et al. Genome-wide analysis of AP2/ERF transcription factors in carrot (Daucus carota L.) reveals evolution and expression profiles under abiotic stress. Mol. Genet. Genom. 290, 2049–2061. https://doi.org/10.1007/s00438-015-1061-3 (2015).
Zhang, H. et al. Arabidopsis AtERF15 positively regulates immunity against Pseudomonas syringae pv. Tomato DC3000 and Botrytis cinerea. Front. Plant Sci. 6, 686–698. https://doi.org/10.3389/fpls.2015.00686 (2015).
Uknes, S. et al. Acquired resistance in Arabidopsis. Plant Cell 4, 645–656. https://doi.org/10.1105/tpc.4.6.645 (1992).
Dahal, D., Pich, A., Braun, H. & Wydra, K. Analysis of cell wall proteins regulated in stems of susceptible and resistant tomato genotypes after inoculation with Ralstonia solanacearum: A proteomics approach. Plant Mol. Biol. 73, 643–658. https://doi.org/10.1007/s11103-010-9646-z (2010).
Acknowledgements
The research was financially supported by China National Tobacco Corporation (Grant No. 110201502018), Hubei tobacco company (Grant No. 027Y2020-002).
Author information
Authors and Affiliations
Contributions
Y.L., L.W., J.F. and Y.Y. wrote the main manuscript text and prepared all figures. All authors participated this research experiments and reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, Y., Wang, L., Sun, G. et al. Digital gene expression analysis of the response to Ralstonia solanacearum between resistant and susceptible tobacco varieties. Sci Rep 11, 3887 (2021). https://doi.org/10.1038/s41598-021-82576-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-82576-8
- Springer Nature Limited
This article is cited by
-
Transcriptome sequencing and expression analysis in peanut reveal the potential mechanism response to Ralstonia solanacearum infection
BMC Plant Biology (2024)
-
Transcriptomic profiling of dynamic alternative splicing during the early response to Ralstonia solanacearum infection in tobacco roots
Plant Growth Regulation (2024)
-
Rhizosphere Microbial Community and Metabolites of Susceptible and Resistant Tobacco Cultivars to Bacterial Wilt
Journal of Microbiology (2023)
-
Transcriptome analysis reveals differential transcription in tomato (Solanum lycopersicum) following inoculation with Ralstonia solanacearum
Scientific Reports (2022)