Abstract
Interleukin-7 (IL-7) is a cytokine known for its importance in T cell development and survival. How IL-7 shapes CD8 T cell responses during an acute viral infection is less understood. We had previously shown that IL-7 signaling deficient mice have reduced accumulation of influenza-specific CD8 T cells following influenza infection. We sought to determine whether IL-7 affects early CD8 T cell expansion in the mediastinal lymph node and effector function in the lungs. Using IL-7Rα signaling deficient mice, we show that IL-7 is required for a normal sized mediastinal lymph node and the early clonal expansion of influenza-specific CD8 T cells therein. We show that IL-7 plays a cell-intrinsic role in the accumulation of NP366–374 and PA224–233-specific CD8 T cells in the lymph node. We also found that IL-7 shapes terminal differentiation, degranulation and cytokine production to a greater extent in PA224–233-specific than NP366–374-specific CD8 T cells. We further demonstrate that IL-7 is induced in the lung tissue by viral infection and we characterize multiple cellular sources that contribute to IL-7 production. Our findings on IL-7 and its effects on lower respiratory diseases will be important for expanding the utility of therapeutics that are currently available.
Similar content being viewed by others
Introduction
The influenza virus is an airway pathogen that infects lung epithelial cells and activates a network of immune cells. It causes seasonal and pandemic outbreaks with major global health and economic impacts. Seasonal variants of influenza can cause death in children, the elderly and immune-compromised individuals1. Vaccination is a cornerstone of the preventative measures taken towards influenza as it arms the adaptive immune system. Multiple cell types in the immune system are required during a response to influenza infection. However, we lack a complete understanding of the cellular aspects and intercellular signaling components that lead to efficient generation of functionally competent immune cells. At the center of immune responses are the cytokine signals that shape various aspects of immune cells2.
A hallmark of our immune response is its ability to develop memory to previously encountered pathogens—T cells are major players in this process. An ideal anti-viral response to influenza and other viruses, requires cytotoxic CD8 T cells for their swift and specific response. CD8 T cells employ multiple methods to kill infected cells and control viral replication, namely, granule-dependent (granzyme B and perforin) and ligand-dependent (Fas-FasL) means3.
In addition to TCR-MHC engagement (signal 1) and co-stimulation (signal 2), cytokine cues (signal 3) have great influence in activating and shaping CD8 T cell responses and their terminal differentiation. Once a CD8 T cell receives these signals, it is driven towards a robust clonal expansion phase whereby a single cell expands to ~ 105 cells4. The signal 3 cytokines that govern T cells are multifaceted and include interleukin-2 (IL-2), IL-6, IL-10, IL-12, IL-15 and others which dictate their terminal differentiation and inflammatory functions5,6.
The common gamma chain (γc) cytokine IL-7 is produced mainly by stromal cells in the bone marrow and thymus. At steady state, it plays an indispensable role in the development of both pre- and pro- B cells and T cells in mice7,8,9,10. IL-7 is important in the development and survival of T cells at specific stages of maturation in the thymus as the expression of IL-7Rα (CD127) is dynamically regulated8,11,12,13. IL-7 shares the IL-7Rα with thymic stromal lymphopoietin (TSLP), an alarmin cytokine that plays a major role in mucosal sites. In addition to its role in development, IL-7 also plays a canonical role in maintenance of memory T cells14. The span of IL-7’s function was further expanded in the past decade when it was shown to be able to shape the effector responses of cytotoxic CD8 T cells by enhancing their responses against tumors15 and bacterial infection14, and reverse T cell exhaustion caused by chronic LCMV infection, thus, preventing liver pathology16. However, the extent to which IL-7 regulates CD8 T cell response to acute viral infections is unknown. We had previously shown that IL-7 but not TSLP is important for the accumulation of influenza-specific CD8 T cells in the lungs but the mechanism by which this occurs is unclear17. Since IL-7 is implicated in over 8 clinical trials for treatment of infections, solid tumors and other chronic conditions, the intricacies of IL-7 signaling in functional outcomes requires further inquiry18.
In this study, we asked: what modulatory effects does IL-7 have on CD8 T cell priming and effector functions during an acute airway influenza infection? We used IL-7Rα knock-in mice (IL-7Rα449F), a hypomorphic model with targeted disruption of a signaling motif that permits development of naïve lymphocytes amenable in infection tests14. We show that in the lung draining mediastinal LNs (mdLNs), IL-7 is important for early priming and accumulation of CD8 T cells specific for influenza NP366–374 and PA224–233 presented on H2Db in a cell intrinsic manner. We also show that IL-7 is important for the terminal differentiation and cytokine production in CD8 T cells. This study will aid in therapeutic development and vaccine adjuvant studies to design combinatorial therapeutic strategies.
Results
IL-7Rα signaling is required for accumulation of influenza-specific CD8 T cells
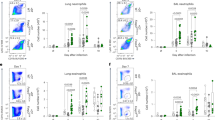
To assess the importance of IL-7 signaling in CD8 T cells following infection with influenza, we used mice expressing a mutated IL-7Rα whereby a cytoplasmic tyrosine important for signal transduction is substituted to a phenylalanine at residue 449 (IL-7Rα449F). We compared WT and IL-7Rα449F mice following infection with A/PR/8/34 (PR8) influenza virus and measured influenza-specific CD8 T cells by flow cytometry using MHC-I tetramers (Suppl. Fig. 1a). We found that IL-7Rα449F mice have reduced proportions of NP366–374 and PA224–233-specific cells within CD8 T cells in the lungs 7 days post-infection (dpi) (Fig. 1a), which phenocopies past observation of this defect at 9 dpi in a new, embryo re-derivation based, specific pathogen free facility17. Since the majority of influenza-specific T cells originate from tissue draining lymph nodes, we examined the mediastinal lymph nodes (mdLNs) of infected mice and found that IL-7Rα449F mice have reduced lymph node sizes, particularly the mdLN, compared to WT mice. Interestingly, unlike WT mice, there was little increase in mdLN size of IL-7Rα449F mice after influenza infection (Fig. 1b). Additionally, enumeration of the total influenza-specific cells in the mdLN at multiple days post infection revealed a consistent and substantial defect that is not due to a delay in expansion kinetics. (Fig. 1c). Consistent with lack of LN hyperplasia, the proportion of influenza-specific cells in IL-7Rα449F mdLN at 5 dpi was reduced indicating a defect in early priming of CD8 T cells (Fig. 1d).
Accumulation of tetramer specific response in IL-7Rα449F is impaired following influenza infection. (a,d) Representative FACS plots and bar graphs of the frequency of NP366–374+ and PA224–233+ cells within CD8 T cells in (a) the lungs 7 dpi and (d) mdLN 5 dpi of WT and IL-7Rα449F mice. Gated within Live B220−, CD8+, (CD44+) cells. (b) Photograph images offering comparison of various mouse lymph nodes and (c) total number of NP366–374+ and PA224–233+ cells in the mdLN of WT and IL-7Rα449F mice at the indicated days post infection. Data are representative of 2–3 experiments with n = 4–7 per genotype. *P < 0.05 as determined by two‐tailed Student's t‐test.
Intrinsic requirement for IL-7Rα signaling in the accumulation of influenza-specific CD8 T cells in the mdLN
Previous reports have shown that IL-7 is required for the generation of lymph nodes independent of the peripheral lymphocyte pool19,20. This likely contributed to the reduced lymph node sizes noted above. Therefore, it is possible that the reduction in influenza-specific CD8 T cell accumulation was due to factors extrinsic to T cells in the lymph node and that IL-7 was indirectly important for shaping the cellular and cytokine environment for optimal T cell activation. To address this, we created bone marrow (BM) chimeric mice whereby we grafted BM cells of wild type (WT) and IL-7Rα449F mice into lethally irradiated RAG-1-deficient hosts (Fig. 2a). Since WT lymphocytes outcompete IL-7Rα449F lymphocytes during development13, we delivered a 1 to 10 ratio of WT to IL-7Rα449F cells respectively. Following engraftment and infection of the hosts, we noted a reversal of this ratio within the CD8 T cell compartment in the mdLN (Fig. 2b). More importantly, IL-7Rα449F CD8 T cells resulted in reduced NP366–374 and PA224–233—specific cells in proportion despite engraftment in a competent niche (Fig. 2b). These data suggest that IL-7Rα signaling plays an intrinsic role necessary for CD8 T cell expansion during influenza infection.
Impairment in tetramer specific response seen in the mdLN of IL-7Rα449F mice is cell intrinsic. (a) Schematic of bone marrow chimera set-up. (b) Representative FACS plots and bar graphs of the frequency of NP366–374+ and PA224–233+ cells within CD8 T cells in the mdLN of WT and IL-7Rα449F chimeric mice 9 dpi. Gated within live, B220−, CD8+, CD44+, CD45.1+ or CD45.2+ cells. Data are representative of two experiments with n = 5–7 per genotype. *P < 0.05 as determined by two‐tailed Student's t‐test.
IL-7Rα plays a role in early priming of CD8 T cells independent of TCR and number of naïve precursors in the mdLN
To determine if the reduction in influenza-specific CD8 T cells was due to reduced numbers of naïve precursors or a result of gaps in TCR repertoire, we adoptively transferred SIINFEKL OVA peptide-specific and MHCI restricted transgenic TCR CD8 T cells from OT-I mice crossed with IL-7Rα449F mice (CD45.2) in to BoyJ mice (CD45.1). We infected these mice with a modified version of the influenza PR8 virus that has the OVA (SIINFEKL) peptide inserted into the stalk of the NA polypeptide (influenza PR8-OVA). Despite delivering equal number (1 × 106) of OT-I and OT-I;IL-7Rα449F CD8 T cells into distinct BoyJ (CD45.1) hosts, OT-I;IL-7Rα449F CD8 T cells did not expand to the same extent as wild type OT-I CD8 T cells 4 dpi (Fig. 3a) in the mdLN. This defect was observed as early as 3 dpi (Suppl. Fig. 1b). Interestingly, the expression of the early activation marker CD5 at 4 dpi was significantly reduced in OT-I;IL-7Rα449F CD8 T cells indicating a defect in priming (Fig. 3b,c). Furthermore, TCR expression on OT-I;IL-7Rα449F CD8 T cells showed a trend towards higher expression at 4 dpi albeit not significantly, further suggesting a possible defect in early priming (Fig. 3b,c).
Expansion of adoptively transferred OTI-IL-7Rα449F CD8 T cells is impaired in the mdLN following influenza infection. (a) Scatter plot and representative bar graph of CD45.2+ Vα2+ CD8 T cells in BoyJ (CD45.1+) mice 4 dpi. Gated within live B220− CD8+ cells. (b) Histogram and (c) bar graph of median florescence intensity (MFI) of activation markers (CD5, TCR (Vα2), and CD69). Data are representative of two experiments. *P < 0.05 and **P < 0.01 as determined by two‐tailed Student's t‐test.
Increased dendritic cell accumulation in the lungs of IL-7Rα449F mice
Evidence of IL-7 cell-intrinsic effects on CD8 T cells does not exclude cell-extrinsic effects. Antigen presenting cells, specifically dendritic cells (DCs) are key to activation of CD8 T cells and their subsequent response. Previous reports have demonstrated that IL-7Rα signaling plays an indirect role in the development of conventional DCs21. Furthermore, IL-7 has been shown to regulate CD4 T cell proliferation in conditions of lymphopenia indirectly though DCs22. We found that in the lungs of IL-7Rα449F mice, CD11b+ CD103− DCs but not CD11b− CD103+ DCs, accumulate up to 9 dpi, while in WT mice these DCs peak at 7 dpi and decrease in numbers at 9 dpi (Suppl. Figs. 1c and 2a). This increased accumulation could be due to cell-extrinsic factors such as increased viral load in the IL-7Rα449F mice as a result of lack of appropriate T cell response. Another plausible reason could be due to impaired lymph node homing signals from chemokines as a result of reduced draining lymph node size. Alternatively, IL-7 may have a direct effect on CD11b+ CD103− DC maturation or migration. To test these hypotheses, we created 50:50 BM chimeras using WT:IL-7Rα449F or WT:WT BM cells (CD45.1:CD45.2) grafted into congenic BoyJ/WT hosts (CD45.1/.2). We found that after infection, WT:IL-7Rα449F ratios of DC subsets were comparable to WT:WT ratios in both the mdLN and lungs for both DC subsets (Suppl. Fig. 2b,c). This suggests that the phenotypic elevation of DCs in IL-7Rα449F mice during influenza infection has a cell extrinsic cause.
IL-7 is inducible in lung tissues in response to influenza
IL-7 is mainly produced by radio-resistant cells such as stromal and epithelial cells of the bone marrow and thymus, where it plays a major role in hematopoiesis and thymopoiesis10,23. A few studies have demonstrated IL-7 expression in various tissues including liver, skin, intestines and lungs24,25,26,27,28. While IL-7 is mainly produced in steady state lungs by lymphatic endothelial cells, its source during inflammation is unclear29,30. To assess IL-7 expression dynamics in response to influenza, we first infected human type II epithelial cells (A549) with influenza and assessed Il7 mRNA using qRT-PCR. We found that IL-7 expression is induced within 24 h following influenza infection correlating with the antiviral response signified by IFN-β and viral replication demonstrated by M1 mRNA transcript expression (Fig. 4a).
IL-7 expression in lung tissues. (a) Quantitative PCR of IL-7, IFN-β and M1 in A549 cells at the indicated times post infection normalized against beta actin. M1 expression is further normalized to time point 0 h. Data is representative of 2 experiments n = 3 per experiment. (b) Expression of IL-7 in various CD45− lung cells using IL-7eGFP/WT mice. Epithelial cells (ECs) are CD45− EpCAM+, stromal cells (SCs) are CD45− EpCAM− CD31− and lymphatic endothelial cells (LECs) are CD45− EpCAM− and CD31+ GP38+. Data are representative of two experiments with n = 4 per genotype. *P < 0.05 as determined by two‐tailed Student's t‐test.
To determine if IL-7 is induced in vivo, we used the IL-7eGFP/WT mice. We noted an increase in the number of cells expressing eGFP at 6 dpi (Fig. 4b). Interestingly, the majority of IL-7-eGFP+ cells during infection are epithelial cells. The number of IL-7-eGFP+ stromal cells also significantly increased during infection. However, IL-7-eGFP+ lymphatic endothelial cells were reduced following infection (Fig. 4b). Altogether, these in vitro and in vivo experiments suggest that lung epithelial cells are responsive to viral infection, and that during influenza infection, they become the primary source of IL-7.
IL-7Rα449F CD8 T cells have reduced terminal differentiation
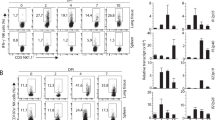
The cytokine milieu that CD8 T cells are exposed to throughout the course of an immune response governs their terminal differentiation to effector cells and hence, their functional capabilities. Among the heterogeneous population of CD8 T cells that emerge during the expansion phase are the T-bethi and granzyme B producing short-lived effector cells (SLECs) that are identified by their expression of the killer cell lectin-like receptor G1 (KLRG1) and low CD127 (IL-7Rα)6,31. Due to extra-physiological expression of the mutated IL-7Rα449F subunit (Suppl. Fig. 3), we limited our use of SLEC markers to KLRG1. Testing the expression of KLRG1 in influenza-specific cells revealed a reduced proportion of KLRG1+ cells in both NP366–374 and PA224–233-specific cells of IL-7Rα449F mice (Fig. 5). However, the difference in KLRG1 expression between WT and IL-7Rα449F mice was greater in PA224–233-specific compared to NP366–374-specific cells (Fig. 5). These data suggest that IL-7Rα signaling plays a role in the terminal differentiation of influenza-specific CD8 T cells.
CD8 T cells of IL-7Rα449F mice have reduced terminal differentiation. Scatter plots and bar graphs showing flow cytometric analysis of KLRG1 expression as a percentage within lung NP366–374 + and PA224–233 + CD8 T cells of WT and IL-7Rα449F mice 7–9 dpi. Data are representative of three experiments with n = 4–6 per genotype. *P < 0.05 as determined by two‐tailed Student's t‐test.
Reduced degranulation and cytokine production by IL-7Rα449F CD8 T cells
Secretion of cytotoxic granules and inflammatory cytokines is a major event in the CD8 T cell effector response. Lysosome associated membrane protein-1 (LAMP-1) or CD107a is a membrane glycoprotein found in the lumen of granzyme B and perforin containing vesicles. Detection of CD107a on the surface of CD8 T cells through flow cytometry after a short stimulation (4 h) with PMA/Ionomycin in the presence of CD107a detecting antibodies provides a direct method for identifying degranulating cells32. Using this method, we noted that CD8 T cells of IL-7Rα449F mice had reduced CD107a expression in antigen experienced cells (CD44+) and in tetramer-stained PA224–233-specific cells indicating decreased degranulation in these populations (Fig. 6a,b). Interestingly, this defect was notable in PA224–233-specific cells but not in NP366–374-specific cells. Furthermore, the proportion of cells expressing CD127 was notably higher in PA224–233-specific cells compared to NP366–374-specific cells in WT mice indicating increased influence of IL-7 on PA224–233-specific cells (Fig. 6c). As mentioned earlier, IL-7Rα449F cells have extra-physiological expression of CD127 (IL-7Rα), however, we found that WT and IL-7Rα449F influenza-specific CD8 T cells experience a similar fold decrease in expression of CD127 compared to naïve CD44− CD8 T cells (Suppl. Fig. 3).
Reduced degranulation of IL-7Rα449F lung CD8 T cells upon re-stimulation. (a,b) CD107a expression as median florescence intensity (MFI) in all antigen experienced (CD44+), NP366–374+ and PA224–233+ CD8 T cells of WT and IL-7Rα449F mice 7 dpi after PMA/Ionomycin re-stimulation. Data presented as (a) FACS plots and (b) bar graphs. Gated within Live B220−, CD8+, CD44+, NP366–374+ or PA224–233 + cells. Data are representative of two experiments with n = 4 per genotype. *P < 0.05 as determined by two‐tailed Student's t‐test. (c) Scatter plots and Bar graphs showing flow cytometric analysis comparison of CD127 expression as a percentage within lung NP366–374 + and PA224–233 + CD8 T cells 7 dpi. Gated within Live B220−, CD8+, CD44+, NP366–374+ or PA224–233+ cells. Data are representative of three experiments with n = 4–5 per genotype. *P < 0.05 as determined by on-way ANOVA.
To determine if IL-7 signaling affects cytokine production, we treated whole lung single cell suspensions from infected mice with NP366–374 and PA224–233 peptides ex vivo and stained intracellular cytokines to detect IFNγ and TNFα using flow cytometry. NP366–374-specific cells generated low proportion of IFNγ+ TNFα+ cells compared to PA224–233-specific cells regardless of mouse genotype (Fig. 7a). However, WT PA224–233-specific cells generated abundant IFNγ+ TNFα+ cells, which were largely absent within IL-7Rα449F PA224–233-specific cells (Fig. 7b). We used TSLPR-/- mice as controls since IL-7Rα is required for both IL-7 and TSLP signaling. We found that TSLPR-/- CD8 T cells presented with reduced cytokine production as well, however, this effect did not follow the same pattern as with IL-7Rα449F mice (Fig. 7a,b).
Deregulated cytokine production in IL-7Rα449F and TSLPR-/- lung CD8 T cells. Representative scatter plots and bar charts of IFN-γ+ TNF-α− or IFN-γ+ TNF-α+ CD8 T cells within (a) NP366–374+ and (b) PA224–233+ CD8 T cells 9 dpi and after peptide (NP or PA) re-stimulation. Representative scatter plots and bar charts of IFN-γ+ TNF-α− or IFN-γ+ TNF-α+ CD8 T cells of IL-7eGFP/eGFP mice within (c) NP366–374+ and (d) PA224–233+ CD8 T cells 9 dpi and after peptide (NP or PA) re-stimulation. Gated within Live B220−, CD8+, tetramer+ cells. Data is representative of three independent experiments with n = 3–5 mice per genotype. *P < 0.05 as determined by one-way ANOVA.
To determine if IL-7 independently affects cytokine production, we used IL-7eGFP/eGFP mice that have an eGFP gene inserted disruptively into an Il7 exon thus serving as an IL-7 ligand knock-out in homozygotes29. Using this mouse model, we established that IL-7 is required for accumulation of IFNγ+ TNFα+ cells within PA224–233-specific but not NP366–374-specific cells (Fig. 7c,d).
IL-7 signaling regulates expression of PD-1 in PA224–233 but not NP366–374-specific CD8 T cells
Expression of inhibitory molecules such as PD-1 is known to be important to negatively regulate T cell activation and limit inflammation by T cells, however, sustained expression of these molecules can lead to dampening of protective immune responses33. To understand how IL-7 affects PA224–233-specific but not NP366–374-specific T cells, we evaluated the expression of the inhibitory receptor PD-1. We showed that IL-7Rα449F and IL-7eGFP/eGFP CD8 T cells have higher expression of this molecule (Fig. 8a,b). Specifically, the increase in PD-1 expression in IL-7Rα449F and IL-7eGFP/eGFP CD8 T cells was only evident in PA224–233-specific cells but not NP366–374-specific cells (Fig. 8a,b). In addition, TSLPR-/- CD8 T cells did not present with increased PD-1 expression (Fig. 8a). Together, this suggests that IL-7 plays distinct roles in CD8 T cell function depending on antigen specificity possibly by regulating PD-1 expression.
Increased PD-1 expression in IL-7 signaling deficient CD8 T cells. Representative histogram plots and bar charts of PD-1 expression in antigen specific lung CD8 T cells of (a) WT versus IL-7Rα449F versus TSLPR-/- and (b) WT versus IL-7eGFP/eGFP. Gated within Live B220−, CD8+, tetramer+ cells. Data is representative of 2–3 independent experiments with n = 3–4 mice per genotype. *P < 0.05 as determined by two‐tailed Student's t‐test.
Discussion
Initial studies of IL-7 have described its role in B-cell lymphopoiesis and thymopoiesis34,35,36. The bone marrow and thymus are the best defined sources of IL-7 production consistent with such roles in primary lymphopoiesis10. Subsequent studies showed a role for IL-7 in memory cell development and maintenance, in effector response to viral infections and in enhancing T cell functions in chronic conditions14,15,16. CD8 T cell expansion and effector function depends on multiple factors including, but not limited to, the cytokine milieu. Previously, we demonstrated that IL-7 is required for the accumulation of tetramer positive CD4 and CD8 T cells during influenza infection17. The mechanism by which IL-7 accomplishes this and its role in other aspects of T cell response have yet to be elucidated.
We addressed these questions by using mice that express a hypomorphic IL-7Rα (IL-7Rα449F) which leads to impaired IL-7 signaling by primarily abrogating STAT5 activation14. This model provides a better alternative to using IL-7Rα-/- mice since IL-7Rα449F mice have defective signaling yet retain sufficient number of T cells to perform infection studies. We have previously used this mouse model to demonstrate an intact CD8 T cell effector response to intracellular Listeria monocytogenes infection14. In the current study, we found that defective IL-7Rα signaling led to reduced accumulation of influenza-specific CD8 T cells in the secondary lymphoid organ (mdLN) at early priming stages (5 dpi) which ultimately led to reduced accumulation of influenza-specific CD8 T cells in the lungs. Examination of IL-7Rα449F mdLN revealed a great reduction in its size. This is consistent with the fact that IL-7 is required for the development of lymphoid tissue inducer (LTi) cells that seed LN anlagens and drive the organogenesis of LNs19. Considering IL-7Rα449F mice had reduced, albeit notable, numbers of influenza-specific CD8 T cells in their lungs, it is unclear where and how these CD8 T cells expand to significant numbers with an abnormal mdLN. It is possible that tertiary lymphoid organs in the lung tissues such as inducible bronchus-associated lymphoid tissue (iBALT) provide a suitable environment for the accumulation of de novo pathogen specific cells without requiring IL-7 or LTi cells37. We have shown that despite such extrinsic factors, IL-7Rα signaling is required cell intrinsically by CD8 T cells for early priming in the mdLN.
It is known that a population of CD8 T cells specific to a distinct peptide do not originate from a single naïve precursor but rather from tens to hundreds of precursors38,39,40. IL-7 signaling deficient mice have reduced thymic output of T cells, and this may result in a more stochastic or reduced chance of a T cell encountering a cognate MHC-peptide leading to reduced clonal expansion. In addition to these effects, IL-7 can play a role cell intrinsically by affecting TCR repertoire via VDJ recombination or TCR sensitivity8,41. We addressed this by adoptive transfer of CD8 T cells bearing a transgenic TCR (OT-I) in equal numbers (WT vs. IL-7Rα449F) intravenously into congenic WT mice. Using this approach, we found that OT-I;IL-7Rα449F CD8 T cells expanded in response to infection with influenza PR8-OVA to a lower extent compared to OT-I CD8 T cells within total host CD8 T cells. Our findings show that IL-7 is intrinsically important for the accumulation of influenza-specific CD8 T cells during early priming phase in the mdLN independent of TCR specificity and the number of naïve T cell precursors. We have previously shown that IL-7Rα449F CD8 T cell form influenza-specific cells normally during systemic in vivo infection with L. monocytogenes yet do not proliferate well when exposed to suboptimal TCR stimulation in vitro in contrast to high dose TCR stimulation14. It is possible a low dose, local influenza infection recapitulates the low level TCR stimulation model whereby IL-7 plays an essential role in CD8 T cells under low TCR avidity activation.
In addition to the intrinsic role that IL-7 plays in CD8 T cells, we found that IL-7Rα449F mice have continued accumulation of CD11b+ DCs in the lungs while in WT mice, the number of CD11b+ DCs peaks at 7 dpi then subsides. Previous studies using IL-7-/- and IL-7Rα-/- mice showed normal development of DC precursors in the BM, however these mice had reduced migratory DCs in secondary lymphoid organs21. Our BM chimera experiments showed that the effect of IL-7 in DC accumulation was indirect. Therefore, the accumulation of DCs we noted in IL-7Rα449F mice was not due to a problem with migration or maturation and was likely due to the fact that viral clearance was impaired which led to continued recruitment of DCs to the lungs.
We have demonstrated for the first time that IL-7 is inducible in lung epithelial cells in response to viral infection in vivo. While the increase in total IL-7-eGFP+ cells in the lung was modest, we noted a shift in the population that are positive for IL-7-eGFP. In naïve mice, the majority of IL-7-eGFP+ cells were lymphatic endothelial cells (LECs) as previously reported29,30. However, following infection with influenza, IL-7-eGFP+ epithelial cells (ECs) and to a lesser extent stromal cells (SCs) expanded while LECs decreased in frequency. Our results demonstrate that IL-7 can be produced by lung tissues and this could shape the function of CD127 expressing CD8 T cells locally. It is unclear to what extent IL-7 produced by epithelial cells influences nearby cells and the significance of the shift in sources of IL-7. In addition, more sensitive approaches are required to compare the levels of IL-7 expression between the different lung tissues.
Terminal differentiation of activated CD8 T cells is important for the generation of short-lived effector cells (SLECs) that express killer cell lectin-like receptor G1 (KLRG1) and low CD1276,31. We found that following infection with influenza, IL-7Rα449F influenza-specific CD8 T cells have reduced expression of KLRG1 and terminal differentiation to SLEC. The difference in KLRG1 expression between WT and IL-7Rα449F mice was greater within PA224–233-specific cells than in NP366–374-specific cells. We also noted a similar trend in CD107a expression between WT and IL-7Rα449F mice whereby the defect in CD107a expression was more pronounced in PA224–233-specific cells than in NP366–374-specific cells. Increased expression of CD127 by PA224–233-specific cells supports the hypothesis that PA224–233-specific cells have elevated dependence on IL-7 signaling. Furthermore, the increased expression of mutated CD127 in IL-7Rα449F mice did not compensate for the loss of Tyr449 signifying the importance of this signaling residue.
Pro-inflammatory cytokines such as IFNγ and TNFα are important during an anti-viral response to help with recruiting and activating other cells. Within NP366–374-specific cells, we did not observe any differences in expression of IFNγ and TNFα between WT and IL-7Rα449F or IL-7eGFP/eGFP CD8 T cells. However, the large reduction in IFNγ+ TNFα+ population in PA224–233-specific cells of IL-7Rα449F or IL-7eGFP/eGFP mice followed a similar pattern to our findings with CD107a. NP366–374-specific cells did not generate IFNγ+ TNFα+ cells as notably as PA224–233-specific cells, consistent with other groups42,43. Our work further corroborates what previous studies have shown, that TSLP shapes effector T cell responses following influenza infection. However, these effects were shown to occur indirectly through programming of DCs44. Altogether, this suggests that IL-7 may differentially regulate the effector function of CD8 T cells based on their antigen-specificity. Further investigation into the downstream signaling is necessary to understand the mechanism that leads to this response.
Previous studies have shown hierarchical differences between NP366–374 and PA224–233-specific CD8 T cells whereby tissue resident memory CD8 T cells that are NP366–374-specific expressed higher levels of inhibitory molecules including PD-1 at 30 dpi and beyond due to persistent antigen exposure and TCR stimulation45. IL-7 is also known to enhance cytokine production and reverse T cell exhaustion by repressing inhibitory pathways during chronic viral infections in mice15,16. It is well established that TCR signaling duration correlates positively with PD-1 expression but little is known about this phenomenon in the context of influenza-specific CD8 T cells46,47. In an acute hepatitis B virus infection, PD-1 expression in CD8 T cells is negatively correlated to CD127 expression, and blocking PD-1 in acute lymphocytic choriomeningitis virus infection increases the frequency of the CD127+ population48,49. Our finding that PD-1 expression is higher within PA224–233-specific but not NP366–374-specific CD8 T cells with IL-7 signaling deficiency is indicative of an antigen-dependent role for IL-7 in regulating their function. This is suggestive of a negative regulatory role for IL-7 on PD-1 expression dependent on TCR specificty. This hypothesis is corroborated by a previous study where infection with a high pathogenicity influenza virus strain, such as PR8, resulted in elevated PD-1 expression in influenza-specific CD8 T cells compared to infection with the low pathogenicity influenza x31 strain43. This in turn inhibited effector function by specifically affecting development of IFNγ+ TNFα+ cells43. Our studies suggest that PA224–233-specific IL-7 signaling deficient CD8 T cells do not receive the necessary signals to down regulate PD-1. Further studies are required to define the relationship between IL-7 and PD-1 in an acute infection setting and the mechanism by which this specifically affects T cells in an antigen-specific manner.
In summary, we have found that IL-7 is required for an optimal response to acute influenza infection as it shapes the early priming stages of CD8 T cells. Moreover, IL-7 produced by lung tissues is important for the terminal differentiation and effector function selectively in antigen-specific CD8 T cells. Various cytokines have the ability to enhance CD8 T cell responses, however, rigorous testing is necessary to evaluate the adverse responses that these cytokines have on bystander cells. Using cytokines such as IL-7 to complement existing therapies may be beneficial given fewer off target effects due to the limited subset of cells that express CD127. IL-7 is currently in clinical trials for treatment of infections and tumors. Additional studies are necessary to expand the use of IL-7 in other conditions and to study its efficacy when delivered in combination with other agents.
Materials and methods
Mice
All mice were housed and used in the Center for Disease Modeling facility (CDM) at the University of British Columbia (UBC) and all work with animals was carried out with approval and in accordance with the ethical guidelines of the University of British Columbia Animal Care and Biosafety Committees. IL‐7Rα449F mice were generated in-house as described14. Briefly, they express a mutant form of the IL-7Rα with a single amino acid mutation from Tyr to Phe at position 449. C57BL/6, BoyJ (B6.SJL‐Ptprca Pepcb/BoyJ) and C57BL/6-Tg (TcraTcrb) 1100Mjb/J (OT-I) mice were obtained from the Jackson Laboratory (Bar Harbour, ME, USA). IL-7eGFP/eGFP mice were a gift from J.M. McCune (UCSF)29. In all cases, age-matched and sex-matched male and female mice between the ages of 6–12 weeks were used.
Virus
Influenza A/PR/8/34 (PR8) was purchased from Charles River Laboratories (Wilmington, MA). Influenza A/PR/8/34-OVA (PR8-OVA, kindly provided by Dr. Tania Watts, University of Toronto) was propagated in-house in chicken eggs as previously described50. Mice were sub-lethally infected under anesthesia (isoflurane) with 5 Hemagglutinin Units (HAU) of influenza PR8 or 64 HAU of influenza PR8-OVA in 12.5 µL of sterile PBS intranasally.
Tissue preparation
Mice were anesthetized with 5% isoflurane in 1 L/min O2 and euthanized by cervical dislocation and perfused with 10 ml cold PBS (+ 5%FBS, 2 mM EDTA). Lungs were excised and processed by mincing with scissors followed by enzymatic digestion using 180 units/ml collagenase IV and 20 µg/ml DNase I (Worthington biochemical LS004188 and LS002139) in 5 ml RPMI incubated at 37 °C for 30–45 min in a shaker incubator before filtering through 70 µm filters and lysing RBCs with ACK lysis buffer. To assess non-hematopoietic cells in the lungs, dispase (1 u/ml) was added to the enzyme cocktail. Mediastinal lymph nodes (mdLN) were collected and crushed through 70 µm filters and suspended as single cells in cold PBS (5%FBS, 2 mM EDTA).
Antibodies and flow cytometry
All cell surface staining was done at 4 °C for 30 min in the dark. Anti-CD8a [53–6.7] (APC-eFluor780), anti-B220 [RA3-6B2] (PE-eFluor610), anti-CD44 [IM7] (PE-Cy7), anti-IFNγ [XMG1.2] (A488), anti-TNFα [MP6-XT22] (PE), anti-MHCII [M5/114.15.2] (FITC), anti-CD11b [M1/70] (PE-Cy7), anti-CD326/EpCAM [G8.8] (PE-Cy7) anti-CD107a [eBio1D4B] (PE) and Rat IgG2a kappa Isotype control [eBR2a] (PE) were purchased from Thermo Fisher (Waltham, Massachusetts). Anti-KLRG1 [MAFA] (APC), anti-CD127 [SB/199] (PE) anti-PD-1 [29F.1A12] (BV510), anti-CD11b [M1/70] (PE-Cy7), anti-CD11c [N418] (biotin), anti-CD45 [30-F11] (Pacific Blue), anti-CD45.2 [104] (PerCP/Cy5.5 or BV421), anti-CD31 [MEC13.3] (biotin), anti-Gp38 [8.1.1] (PE) and anti-F4/80 [BM8] (PE-Cy7) were purchased from Biolegend (San Diego, California). Anti-CD103 [M290] (PE), anti-TCR Vα2 [B20.1] (PE) and anti-CD45.1 [A20] (APC) were purchased from BD Biosciences (Franklin Lakes, New Jersey). Anti-CD11c [N418] (Alexa fluor-647), anti-CD45.1 [A20] (A488), anti-B220 [RA-6B2] (FITC) and anti-F4/80 [BM8] (biotin) were purchased from AbLab (Vancouver, British Columbia).
Tetramer staining was done at room temperature for 30 min in the dark. H2-Kb tetramers loaded with immune-dominant NP366–374 and PA224–233 peptides from influenza and labeled with Brilliant Violet-421 or Alexa fluor-647 were manufactured and donated by the NIH Tetramer Core Facility (Atlanta, GA).
Viability staining [cat# L34957 and 65-0865-14] (Thermo Fisher) was used according to manufacturer’s instructions.
Samples were collected on either a FACSCanto, LSRII (BD Biosciences) or the Attune NxT (Thermo Fisher) and data were analyzed with FlowJo software Tree Star (Ashland, Oregon).
Bone marrow chimeras
Recipient mice were irradiated with 2 doses of 6.5 grey (Gy) or 650 rad at least 4 h apart. For the following 10 days, they were supplemented with antibiotics ad libitum (2 mg/ml neomycin sulfate). 24 h after radiation, femurs and tibias were collected from donor CD45.1 and CD45.2 mice (WT and IL-7Rα449F respectively). RBCs were removed using sterile ACK lysis. For tetramer response experiments, a total of 1 × 106 donor bone marrow (BM) cells were injected intravenously (I.V.) at a 1:1 ratio to deliver WT:WT or 1:10 ratio to deliver WT:IL‐7Rα449F into Rag1-/- hosts. For dendritic cell experiments, a total of 5 × 105 donor BM cells were injected I.V. at a 1:1 ratio to deliver WT:WT or WT: IL‐7Rα449F into C57BL/6 J;Boy/J (CD45.1/0.2) hosts. 6–8 weeks elapsed for reconstitution before challenge with influenza infection. After euthanasia, spleens and BMs were assessed for reconstitution efficiency and ratios.
Adoptive transfer
Single cell suspensions were prepared from multiple OT-I and OT-I;IL-7Rα449F mice spleens and CD8 T cells were purified using the CD8 T cell negative selection kit (EasySep™ Mouse CD8 + T Cell Isolation Kit) from Stem Cell Technologies. 1 × 106 cells were transferred I.V. into BoyJ (CD45.1) hosts and 24 h later hosts are challenged with 64HAU PR8-OVA intranasally. MdLN was harvested at experimental endpoint and cells were stained for surface markers and analyzed by flow cytometry as described above.
Cell culture and Il-7 RT-qPCR
A549 (ATCC CCL-185) human type II alveolar epithelial cells were obtained from the American Type Culture Collection (ATCC) (Manassas, Virginia). Cells were passaged and expanded in 10% FBS F-12 K Medium from ATCC (Cat No. 30-2004). For experimental use, 5 × 105 A549 cells were seeded into 6-well plates in media and expanded for 24 h to achieve confluence. After 1 h of serum starvation, cells were infected with 200 HAU PR8 in PBS and incubated for 1 h on a plate shaker to initiate infection. Virus containing PBS was aspirated and replaced with F-12 K media containing 0.5% BSA and 0.5 µg/mL N-tosyl-L-phenylalanine chloromethyl ketone (TPCK) treated trypsin. Cells were then incubated for the assigned experimental time points. Cells were lysed and RNA was extracted using PureLink RNA Mini Kit (Thermo Fisher). After treatment with amplification grade DNase I (Thermo Fisher), cDNA was generated using the iScript cDNA synthesis kit (Bio-Rad) and cDNA quantification was performed using the Ssofast EvaGreen Supermix kit (Bio-Rad). Primer sequences are as follows. β-actin Forward: GAC ATG GAG AAA ATC TG; β-actin Reverse: ATG ATC TGG GTC ATC TTC TC; Human IL-7 Forward: CCA GGT TAA AGG AAG AAA ACC; Human IL-7 Reverse: TTT CAG TGT TCT TTA GTG CC; Human IFN-β Forward: ACG CCG CAT TGA CCA TCT AT; Human IFN-β Reverse: GTC TCA TTC CAG CCA GTG CTA; M1 Forward: AGA TGA GTC TTC TAA CCG AGG TCG; M1 Reverse: TGC AAA AAC ATC TTC AAG TCT CTG. Measurements were acquired using the CFX96 Touch Real-Time PCR Detection System (Bio-Rad).
Ex vivo T cell re-stimulation
Lungs from mice infected (7–9 days) with PR8 were excised and prepared as above. To measure CD107a, 5 × 106 lung cells were re-stimulated for 4 h (37 °C, 5% CO2) with 1% BSA RPMI containing 50 ng/ml PMA and 500 ng/ml Ionomycin from Sigma-Aldrich (St. Louis, Missouri); Monensin from BD Biosciences (Franklin Lakes, New Jersey) used according to manufacturer’s instructions; and anti-CD107a [eBio1D4B] PE32. Following re-stimulation, cells were stained for viability and surface markers then analyzed by flow cytometry.
To measure IFNγ and TNFα, 5 × 106 lung cells were re-stimulated for 3 h (37 °C, 5% CO2) with 10 nM NP366–374 and PA224–233 peptides from Anaspec (Fremont, California) in the presence of Brefeldin/A from BD Biosciences (Franklin Lakes, New Jersey) in 1% BSA RPMI. Following re-stimulation, cells were stained for viability and surface markers followed by intracellular cytokine staining using the Cytofix/Cytoperm kit from BD Biosciences (Franklin Lakes, New Jersey) then analyzed by flow cytometry.
Statistical analysis
All analyses were performed on GraphPad Prism 8 (La Jolla, CA, USA). Samples that were identified as outliers by the Gibbs tests were excluded from the analysis. Two-tailed t-tests and one-way ANOVA with Tukey’s post-test were performed as appropriate and P<0.05 was considered statistically significant. Unless specified, all error analysis in bar charts are presented as S.E.M.
Ethics
All experiments described using mice had ethical and procedural approval from the University of British Columbia Animal Care Committee (protocol A17-0249) as per guidelines set by the Canadian Council on Animal Care. No human studies were undertaken. All methods are reported in accordance with ARRIVE guidelines.
References
Horimoto, T. & Kawaoka, Y. Influenza: Lessons from past pandemics, warnings from current incidents. Nat. Rev. Micro. 3(8), 591–600 (2005).
Schotsaert, M., Saelens, X. & Leroux-Roels, G. Influenza vaccines: T-cell responses deserve more attention. Expert Rev. Vaccines 11(8), 949–962 (2012).
Trapani, J. A. & Smyth, M. J. Functional significance of the perforin/granzyme cell death pathway. Nat. Rev. Immunol. 2(10), 735–747 (2002).
Buchholz, V. R., Schumacher, T. N. & Busch, D. H. T cell fate at the single-cell level. Annu. Rev. Immunol. 34(1), 65–92 (2016).
Valbon, S. F., Condotta, S. A. & Richer, M. J. Regulation of effector and memory CD8(+) T cell function by inflammatory cytokines. Cytokine 82, 16–23 (2016).
Joshi, N. S. et al. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity 27(2), 281–295 (2007).
Patton, D. T., Plumb, A. W. & Abraham, N. The survival and differentiation of pro-B and pre-B cells in the bone marrow is dependent on IL-7Rα Tyr449. J. Immunol. 193(7), 3446–3455 (2014).
Boudil, A. et al. IL-7 coordinates proliferation, differentiation and Tcra recombination during thymocyte beta-selection. Nat. Immunol. 16(4), 397–405 (2015).
Hara, T. et al. Identification of IL-7-producing cells in primary and secondary lymphoid organs using IL-7-GFP knock-in mice. J. Immunol. 189(4), 1577–1584 (2012).
Mazzucchelli, R. I. et al. Visualization and identification of IL-7 producing cells in reporter mice. PLoS ONE 4(11), e7637 (2009).
Magri, M. et al. Notch ligands potentiate IL-7-driven proliferation and survival of human thymocyte precursors. Eur. J. Immunol. 39(5), 1231–1240 (2009).
Munitic, I. et al. Dynamic regulation of IL-7 receptor expression is required for normal thymopoiesis. Blood 104(13), 4165–4172 (2004).
Plumb, A. W. et al. Interleukin-7 in the transition of bone marrow progenitors to the thymus. Immunol. Cell Biol. 95(10), 916–924 (2017).
Osborne, L. C. et al. Impaired CD8 T cell memory and CD4 T cell primary responses in IL-7R alpha mutant mice. J. Exp. Med. 204(3), 619–631 (2007).
Pellegrini, M. et al. Adjuvant IL-7 antagonizes multiple cellular and molecular inhibitory networks to enhance immunotherapies. Nat. Med. 15(5), 528–536 (2009).
Pellegrini, M. et al. IL-7 engages multiple mechanisms to overcome chronic viral infection and limit organ pathology. Cell 144(4), 601–613 (2011).
Plumb, A. W. et al. Interleukin-7, but not thymic stromal lymphopoietin, plays a key role in the T cell response to influenza a virus. PLoS ONE 7(11), e50199 (2012).
Buckley, R. H. Molecular defects in human severe combined immunodeficiency and approaches to immune reconstitution. Annu. Rev. Immunol. 22(1), 625–655 (2004).
Chappaz, S. & Finke, D. The IL-7 signaling pathway regulates lymph node development independent of peripheral lymphocytes. J. Immunol. 184(7), 3562–3569 (2010).
Schmutz, S. et al. Cutting edge: IL-7 regulates the peripheral pool of adult ROR gamma+ lymphoid tissue inducer cells. J. Immunol. 183(4), 2217–2221 (2009).
Vogt, T. K., Link, A., Perrin, J., Finke, D. & Luther, S. A. Novel function for interleukin-7 in dendritic cell development. Blood 113(17), 3961–3968 (2009).
Guimond, M. et al. Interleukin 7 signaling in dendritic cells regulates the homeostatic proliferation and niche size of CD4+ T cells. Nat. Immunol. 10(2), 149–157 (2009).
Martin, C. E. et al. Interleukin-7 availability is maintained by a hematopoietic cytokine sink comprising innate lymphoid cells and T cells. Immunity 47(1), 171–82 e4 (2017).
Sawa, Y. et al. Hepatic interleukin-7 expression regulates T cell responses. Immunity 30(3), 447–457 (2009).
Adachi, T. et al. Hair follicle-derived IL-7 and IL-15 mediate skin-resident memory T cell homeostasis and lymphoma. Nat. Med. 21, 1272–1279 (2015).
Watanabe, M. et al. Interleukin 7 transgenic mice develop chronic colitis with decreased interleukin 7 protein accumulation in the colonic mucosa. J. Exp. Med. 187(3), 389–402 (1998).
Shalapour, S. et al. Commensal microflora and interferon-gamma promote steady-state interleukin-7 production in vivo. Eur. J. Immunol. 40(9), 2391–2400 (2010).
Jin, J. O. & Yu, Q. Systemic administration of TLR3 agonist induces IL-7 expression and IL-7-dependent CXCR3 ligand production in the lung. J. Leukoc. Biol. 93(3), 413–425 (2013).
Miller, C. N. et al. IL-7 production in murine lymphatic endothelial cells and induction in the setting of peripheral lymphopenia. Int. Immunol. 25(8), 471–483 (2013).
Dahlgren, M. W. et al. Adventitial stromal cells define group 2 innate lymphoid cell tissue niches. Immunity 50(3), 707–22 e6 (2019).
Remmerswaal, E. B. M. et al. Expression of IL-7Ralpha and KLRG1 defines functionally distinct CD8(+) T-cell populations in humans. Eur. J. Immunol. 49(5), 694–708 (2019).
Betts, M. R. & Koup, R. A. Detection of T-Cell Degranulation: CD107a and b. Methods in Cell Biology Vol. 75, 497–512 (Academic Press, 2004).
Sharpe, A. H. & Pauken, K. E. The diverse functions of the PD1 inhibitory pathway. Nat. Rev. Immunol. 18(3), 153–167 (2018).
Goodwin, R. G. et al. Human interleukin 7: Molecular cloning and growth factor activity on human and murine B-lineage cells. Proc. Natl. Acad. Sci. U S A. 86(1), 302–306 (1989).
Namen, A. E. et al. Stimulation of B-cell progenitors by cloned murine interleukin-7. Nature 333(6173), 571–573 (1988).
Murray, R., Suda, T., Wrighton, N., Lee, F. & Zlotnik, A. IL-7 is a growth and maintenance factor for mature and immature thymocyte subsets. Int. Immunol. 1(5), 526–531 (1989).
Rangel-Moreno, J. et al. The development of inducible bronchus-associated lymphoid tissue depends on IL-17. Nat. Immunol. 12(7), 639–646 (2011).
Jenkins, M. K. & Moon, J. J. The role of naive T cell precursor frequency and recruitment in dictating immune response magnitude. J. Immunol. 188(9), 4135–4140 (2012).
Arstila, T. P. et al. A direct estimate of the human alphabeta T cell receptor diversity. Science 286(5441), 958–961 (1999).
Casrouge, A. et al. Size estimate of the alpha beta TCR repertoire of naive mouse splenocytes. J. Immunol. 164(11), 5782–5787 (2000).
Deshpande, P. et al. IL-7- and IL-15-mediated TCR sensitization enables T cell responses to self-antigens. J. Immunol. 190(4), 1416–1423 (2013).
Belz, G. T., Xie, W. & Doherty, P. C. Diversity of epitope and cytokine profiles for primary and secondary influenza a virus-specific CD8+ T cell responses. J. Immunol. 166(7), 4627–4633 (2001).
Rutigliano, J. A. et al. Highly pathological influenza A virus infection is associated with augmented expression of PD-1 by functionally compromised virus-specific CD8+ T cells. J. Virol. 88(3), 1636–1651 (2014).
Yadava, K. et al. TSLP promotes influenza-specific CD8+ T-cell responses by augmenting local inflammatory dendritic cell function. Mucosal. Immunol. 6(1), 83–92 (2013).
Wang, Z. et al. PD-1(hi) CD8(+) resident memory T cells balance immunity and fibrotic sequelae. Sci. Immunol. 4(36), eaaw1217 (2019).
Bally, A. P., Austin, J. W. & Boss, J. M. Genetic and epigenetic regulation of PD-1 expression. J. Immunol. 196(6), 2431–2437 (2016).
Youngblood, B. et al. Chronic virus infection enforces demethylation of the locus that encodes PD-1 in antigen-specific CD8(+) T cells. Immunity 35(3), 400–412 (2011).
Boettler, T. et al. Expression of the interleukin-7 receptor alpha chain (CD127) on virus-specific CD8+ T cells identifies functionally and phenotypically defined memory T cells during acute resolving hepatitis B virus infection. J. Virol. 80(7), 3532–3540 (2006).
Ahn, E. et al. Role of PD-1 during effector CD8 T cell differentiation. Proc. Natl. Acad. Sci. U S A. 115(18), 4749–4754 (2018).
Jenkins, M. R., Webby, R., Doherty, P. C. & Turner, S. J. Addition of a prominent epitope affects influenza A virus-specific CD8+ T cell immunodominance hierarchies when antigen is limiting. J. Immunol. 177(5), 2917–2925 (2006).
Acknowledgements
We would like to thank Dr. Adam Plumb, Julia Lu, Etienne Melese and Dr. Lisa Osborne for critical revision of this manuscript. We would also like to thank the staff at the Centre for Disease Modeling for animal husbandry and the UBC Flow Cytometry Facility. Work in the laboratory was supported by a grant to NA from the Canadian Institutes of Health Research (MOP-126060 and PJT-156119). AS was supported by the John Richard Turner Fellowship.
Author information
Authors and Affiliations
Contributions
A.S. conceived and designed the project, performed and analyzed experiments, and wrote the manuscript. J.J., H.B.S. and C.Y. performed and analyzed experiments and reviewed the manuscript. J.S. performed and analyzed experiments. N.A. conceived and designed the project and reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sheikh, A., Jackson, J., Shim, H.B. et al. Selective dependence on IL-7 for antigen-specific CD8 T cell responses during airway influenza infection. Sci Rep 12, 135 (2022). https://doi.org/10.1038/s41598-021-03936-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-03936-y
- Springer Nature Limited