Abstract
Genetic diversity feeds the evolutionary process and allows populations to adapt to environmental changes. However, we still lack a thorough understanding of why hotspots of genetic diversity are so 'hot'. Here, we analysed the relative contribution of bioclimatic stability and genetic admixture between divergent lineages in shaping spatial patterns of genetic diversity in the common toad Bufo bufo along the Italian peninsula. We combined population genetic, phylogeographic and species distribution modelling (SDM) approaches to map ancestral areas, glacial refugia, and secondary contact zones. We consistently identified three phylogeographic lineages, distributed in northern, central and southern Italy. These lineages expanded from their ancestral areas and established secondary contact zones, before the last interglacial. SDM identified widespread glacial refugia in peninsular Italy, sometimes located under the present-day sea-level. Generalized linear models indicated genetic admixture as the only significant predictor of the levels of population genetic diversity. Our results show that glacial refugia contributed to preserving both levels and patterns of genetic diversity across glacial-interglacial cycles, but not to their formation, and highlight a general principle emerging in Mediterranean species: higher levels of genetic diversity mark populations with substantial contributions from multiple genetic lineages, irrespective of the location of glacial refugia.
Similar content being viewed by others
Introduction
Intraspecific genetic variation feeds the evolutionary process and affects biodiversity patterns at all levels of biological organization. It provides populations with the potential to adapt to changes in their biotic and abiotic environment1,2, as confirmed in studies of experimental evolution3. Levels of genetic diversity have been associated to the extinction risk, and have been estimated to be 30% lower in threatened species than in their non-threatened relatives4,5. Moreover, genetic diversity within populations can drive ecological dynamics shaping biological communities and ecosystem functions (see6 for a review). For instance, some recent studies7,8 found significant positive effects of genetic diversity in plant populations on species richness, abundance, and productivity of associated biological communities, with important implications in applied sciences, such as ecological restoration9.
Given the importance of genetic diversity within populations, the analysis and interpretation of spatial patterns of variation across species ranges have been a long-lasting endeavour in evolutionary biology10. A major research arena has involved the identification of hotspots of intraspecific genetic diversity, that is, geographic regions harbouring exceptionally high diversity11,12. These hotspots are increasingly recognised as key targets in conservation biology11, 13,14, and their correct identification is an important step in designing effective strategies for the long-term persistence of populations in the face of global change15.
Despite their theoretical and applied importance, we still lack a thorough understanding of why hotspots of genetic diversity are so 'hot'. In more than thirty years of phylogeographic and population genetic investigations, hotspots of genetic diversity have often been observed in close geographic association with major Pleistocene glacial refugia. For example, southern European peninsulas, south-western and south-eastern North America, and tropical Australia have been identified as major glacial refugia as well as hotspots of biodiversity, at both species and intraspecific levels16. This widespread geographic association between refugia and hotspots of biodiversity has classically been viewed as evidence for a causal link between prolonged bioclimatic stability and high levels of intraspecific genetic diversity17. Additionally, hotspots may also result from secondary contact and admixture between intraspecific lineages, differentiated within sub-refugia during periods of unfavourable climatic conditions17. Under the latter scenario, hotspots of genetic diversity would in fact be melting-pots18,19. Although they were originally treated as alternative11, prolonged bioclimatic stability and secondary contact and admixture are not mutually exclusive scenarios for the formation of hotspots of intraspecific genetic diversity. In fact, substantial evidence has been gathered in favour of each scenario (e.g.20,21,22) with different levels of genetic diversity that can be explained for different populations by two factors: distance from putative refugia and extent of admixture23,24. However, the relative contribution of the two factors to the formation of spatial patterns of genetic variation, and particularly of hotspots, remains poorly explored (but see11,18,19,20,21,22).
Here, we address this question by analysing the contribution of bioclimatic stability and admixture following secondary contact in shaping the fine-scale spatial patterns of genetic diversity in the common toad Bufo bufo in peninsular Italy, a major hotspot of genetic diversity for the species25,26. The common toad is widely distributed in temperate habitats of the western Palearctic and range-wide phylogeographic studies have identified three divergent mtDNA lineages of the common toad in the Italian peninsula25,26,27. One lineage is restricted to southern Italy and Sicily, a second one ranges from central to north-central Italy and a third one is distributed in northern Italy and neighbouring areas. In this study we aimed to dissect the spatial patterns of genetic variation of B. bufo populations in its Pleistocene Italian refugium. We combined population genetic and phylogeographic tools with species distribution modelling, in order to (i) assess the fine-scale population genetic structure and diversity of B. bufo in the Italian peninsula; (ii) infer the location of ancestral areas, glacial refugia and secondary contact zones, and (iii) investigate the relative contribution of prolonged habitat stability and admixture between lineages in the formation and evolution of the Italian hotspot of genetic diversity for the species.
Results
Phylogeographic analyses based on mtDNA
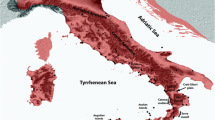
We sequenced two mitochondrial DNA gene fragments in 231 Bufo bufo individuals from 70 sampling localities: a 722-bp fragment of the Cytochrome B gene (CytB) and a 517-bp fragment of the mitochondrial 16 s rRNA gene. In the combined mtDNA dataset (1239 bp), we found 83 different haplotypes defined by 160 variable positions. Two haplotypes found in nine individuals from three localities (13, 14 and 18) were identified as belonging to Bufo spinosus (Fig. 1 and Table 1).
Maximum likelihood phylogenetic network of the Bufo bufo mitochondrial haplotypes found in Italy, and geographic distribution of the main haplotype groups. Circle sizes are proportional to haplotype frequency, and black dots represents missing intermediate haplotypes. Populations are numbered as in Table 1. The map was drawn using the software Canvas 11 (ACD Systems of America, Inc.).
The phylogenetic network outlines four main B. bufo haplogroups, with a clear geographic structure (Fig. 1): a north-eastern haplogroup, spanning from the eastern and central Alps to the northern side of the northern Apennines, a north-western haplogroup, restricted to the western Alps and the Provence, a central haplogroup, spanning from the northern to the central portion of the Apennine chain and a southern haplogroup, spanning from the central Apennines to the southernmost peninsular populations, including those in Sicily. The southern and central haplogroups co-occurred in the geographically intermediate localities 45, 51 and 53, whereas the north-western and central haplogroups co-occurred in a single locality (24). Haplotypes of B. spinosus and B. bufo were found co-occurring in the Provence (locality 13) and in the Ligurian Alps (locality 18).
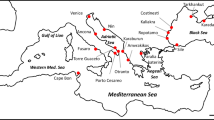
Bayesian phylogeographic analyses were conducted separately for the four main B. bufo mtDNA lineages. For each lineage, runs converged to a stationary distribution and had satisfactory Effective Sample Size (ESS) values (> 200). The ancestral areas of the four lineages were mapped in distant regions along the Italian peninsula (Fig. 2). The ancestral areas of the north-eastern and the north-western lineages were most likely positioned within the Ligurian Alps (time to the most recent common ancestor—TMRCA—median estimate: 418 ky, 95% HPD: 157–845 ky), and close to the Venetian Prealps (TMRCA: 416 ky, 95%HPD: 190–775 ky), respectively. The central lineage had its ancestral area projected within lowlands close to the Apennines (TMRCA: 511 ky, 95%HPD: 255–872 ky), whereas the ancestral area of the southern lineage likely occurred along the mountain massifs in the Calabria region (TMRCA: 498 ky, 95%HPD: 249–859 ky). Spatial diffusion processes from the ancestral areas likely occurred earlier for the southern and central lineages than for the two northern lineages (see Supplementary File S1). However, all the inferred events of range expansion and secondary contact among lineages occurred well before the last glacial maximum (LGM). Based on the temporal patterns of spatial diffusion, the secondary contact between the southern and the central lineages also pre-dates the last interglacial (120–140 ky), whereas the secondary contact between the central and the north-eastern lineage likely occurred close to the last interglacial.
Ancestral areas of the genetic lineages of Bufo bufo at their respective time to the most recent common ancestor (TMRCA), as estimated by the Bayesian phylogeographical analyses. Polygons represent 10% to 70% highest posterior density (HPD) regions of the geographical locations of the ancestral areas. The map was drawn using the software Canvas 11 (ACD Systems of America, Inc.); photo: Bufo bufo (from https://www.dreamstime.com).
Population structure and genetic diversity based on microsatellite data
A total of 500 individuals from 57 populations was genotyped at nine microsatellite loci, with 3.8% of missing data. Locus Bspi 3.02 was removed from the dataset because it tested positive for null alleles in fourteen populations. There were no significant deviations from Hardy–Weinberg or linkage equilibria after applying the Bonferroni correction for multiple tests. Across all populations, the number of alleles per locus ranged from four (Bspi 3.11) to 54 (Bspi 4.29). Allelic richness and mean expected heterozygosity estimates for each population are presented in Table 1. Population 51 (Central Italy) showed the highest values of genetic diversity, whereas the lowest values of heterozygosity and allelic richness were observed in several samples from the southernmost section of the Italian peninsula and Sicily.
TESS analyses revealed strong genetic structure in Italian B. bufo for the nuclear markers. The plots of values for the deviance information criterion (DIC) versus the number of clusters (K) reached a plateau at K = 4, indicating that four main genetic clusters occur in the study area (Fig. 3). The spatial distribution of these clusters shows a strong geographical signal: one cluster is widespread from the Alps to the northern side of the northern Apennines; a second cluster spans from the northern to the central Apennines; a third one spans from the central to the southern Apennines and Sicily and a fourth cluster is restricted to the Provence and to the westernmost Ligurian populations. This cluster is strongly differentiated from the other clusters and represents populations of B. spinosus (see also27,28). Bar-plots showing individual admixture proportions and pie-charts showing the average proportion of each cluster in each sampled population are presented in Fig. 3. We found widespread admixture between the southern and central genetic clusters, encompassing most of the Apennine chain. Evidence for admixture was also observed in northern populations, between the central and northern clusters. In contrast, evidence for hybridisation and admixture between B. bufo and B. spinosus was geographically restricted to their area of close proximity in the Ligurian Alps.
Genetic structure of the Italian Bufo bufo populations, as inferred by the Bayesian clustering analysis implemented in TESS based on eight microsatellite loci. The bar plot on the left shows the admixture proportions of each individual for the four genetic clusters identified; the pie diagrams on the maps show the frequency of each cluster within the studied populations. Populations are numbered as in Table 1. The line chart shows values of the deviance information criterion (DIC) statistics estimated for models with the number of genetic clusters (K) ranging from 2 to 10. The map was drawn using the software Canvas 11 (ACD Systems of America, Inc.).
Identification of glacial refugia
We obtained 581 species occurrences for the central lineage and 256 for the southern lineage (Supplementary Table S2), reduced by the thinning procedure to 369 and 154 occurrences, respectively. A total of 6655 background points for the central lineage and 4715 for the southern lineage were used in the calibration procedure.
The variance inflation factor (VIF) analysis selected seven variables for the central lineage and six variables for the southern lineage (see Supplementary Table S3). All models showed a good performance: mean AUC was 0.79 (SD = 0.02) for the central lineage and 0.81 (SD = 0.05) for the southern lineage. For both lineages, GBM was the model with the highest AUC values and GLM the one with the lowest. Model performance indices are given in Supplementary Table S3.
The current SDM is consistent with previous knowledge about the distribution of the common toad29: high suitability areas are scattered throughout most of the Italian peninsula, where the species is more common at medium elevations and rare and isolated in the main plains and at higher elevations (see Supplementary Figure S1). The putative climatic refugia for the southern lineage at the LGM (Fig. 4 and Supplementary Figure S2) covered most of the Calabrian and Apulian coastlines (predicted by all GCMs), as well as the inland of the same areas (predicted by two GCMs). The putative glacial refugia for the central lineage only covered a small area along the Tyrrhenian coastline in Tuscany (predicted by two GCMs).
Putative Pleistocene glacial refugia for the central (A) and southern (B) lineages of Bufo bufo, as inferred by the species distribution modelling (SDM) calibrated under current bioclimatic conditions and projected at the last glacial maximum (21 kya). Orange: areas of at least two out of three general circulation models (GCMs); red: areas of concordance among all the GCMs; black: areas of “stability”, defined as regions of overlap between current, glacial (two out of three GCMs) and ancestral (Bayesian phylogeographic analysis) areas of species presence. The map was drawn using the software Canvas 11 (ACD Systems of America, Inc.).
Predictors of genetic diversity
Results from the generalised linear models (GLMs) are summarised in Table 2. Irrespective of the genetic diversity index used as dependent variable, admixture between divergent lineages was the only significant predictor of population genetic diversity. On the contrary, refugia and areas of stability showed no significant effects on genetic diversity.
Discussion
In line with previous studies25,26,28, mitochondrial and microsatellite markers consistently identified three main lineages of Bufo bufo, ranging in southern, central and northern Italy. The phylogeographic boundaries between lineages, although sharper with mtDNA than with the microsatellite markers, coincide with two well-known suture zones of the Italian peninsula: the lower Volturno-Calore river basin and the northern Apennines12. The extent of phylogeographic concordance with previously studied organisms from the same area is substantial, in terms of the co-distribution of intraspecific lineages20,30,31,32,33,34,35,36,37. However, the spatial pattern of genetic variation observed within B. bufo populations shows some remarkable features, that may well exemplify the disparity between co-distribution of phylogeographic lineages and correlated population histories.
The geographic distribution of the southern lineage is bounded by the island of Sicily to the south and the lower Volturno-Calore river basin in central Italy to the north. This distribution closely matches phylogeographic patterns in co-distributed taxa (e.g.20,30,31,32,33,34). However, the spatial distribution of genetic variation within the range of the southern lineage of B. bufo contrasts with that found in other species studied with enough sampling depth, typically comprising finer population structure and multiple population units arranged along the south-north axis18,20,31,34,35,36,37,38. The south of the Italian peninsula has been recurrently identified as a hotspot of intraspecific genetic diversity14,18,31. Conversely, neither mtDNA nor microsatellites provided evidence for further population structure within the southern lineage of B. bufo, and, accordingly, the southernmost area of the peninsula is rather a ‘cold spot’ of genetic diversity for this species (Table 1). Throughout the Plio-Pleistocene, the southern portion of the Italian peninsula was repeatedly fragmented by glacio-eustatic marine transgressions39. These cycles of fragmentation into paleo-islands followed by their re-assembly, have had a major impact on the evolution of the regional temperate biota, likely including B. bufo populations. The lower Volturno-Calore river basin was precisely the northernmost site affected by repeated marine transgressions, which plausibly played a role in the divergence between the southern and the central lineages of B. bufo (estimated at 1.7 My ago26). The absence of population structure of the extant B. bufo lineage in southern Italy may result from two processes. Either the range expansion of the extant southern lineage swamped all other pre-existing population units, or these went extinct before this range expansion (see Supplementary Figure S3 and Supplementary File S1). In the former case, however, admixture among divergent population units during the expansion should have inflated genetic diversity in southern Italy, at least in the nuclear genome, as observed in other taxa18,31. Thus, the minimal level of genetic diversity observed in this area fits better with a scenario of early disappearance of—probably small and insular—population units prior to the most recent expansion event in southern Italy and Sicily.
The geographic distribution of the central lineage parallels the distribution of intraspecific lineages in other organisms, including reptiles34 and amphibians26,27,28,29,30,31,32,33,34,35,36,37, as well as several cryptic endemic species32. In this case, however, concordance also involves the spatial and temporal features of phylogeographic reconstructions. For example, Triturus carnifex35 and Hyla intermedia36 have intraspecific lineages (i) co-distributed with the central lineage of B. bufo, (ii) with closest affinities with lineages in southern Italy, and (iii) not showing evidence of further population sub-structure within this area. Whether these multiple lines of phylogeographic concordance are genuine realisations of correlated population histories in response to paleoenvironmental changes requires further research.
At first glance, the geographic distribution of the northern lineage, albeit incompletely captured by our sampling design (see27), appears in concordance with the distribution of several phylogeographic lineages sandwiched between the Alps and the northern Apennines34,35,36,44. However, in B. bufo this probably involved a unique biogeographic route, encompassing two distinct colonisation events of the river Po plain, one from the east and one from the west. Indeed, this lineage is characterised by a single nuclear gene pool (see also28), but two distinct mtDNA sub-lineages, with western and central—eastern European distributions, respectively, coincident with the e2 and e3 lineages of25. Arntzen et al.27 suggested that the two colonisation events occurred after the LGM, eastward from a western refugium, and westward from the northern Balkans. Our results support this scenario, with some amendment. We modelled two distinct ancestral areas for the two mtDNA sub-clades, and we located one area in the Ligurian Alps, and one area close to the Venetian Prealps, respectively (Fig. 2). However, our Bayesian phylogeographic reconstructions (Supplementary File S1) showed that the eastern sub-clade completed most of its range expansion before the last glacial cycle, whereas the western sub-clade completed most of its range expansion during the last glaciation. Finally, the absence of two sub-groups of B. bufo in northern Italy in the nuclear dataset, and the occurrence of B. spinosus in the putative range of the e2 sub-clade (Fig. 3) might be explained by a late expansion of B. spinosus in the north-west, which then hybridised with local B. bufo populations picking up the e2 mtDNA lineage in the process27,28.
Our correlative models indicate that the distance from glacial refugia or stability areas do not contribute much to explaining levels of genetic diversity within B. bufo populations in peninsular Italy, irrespective of the genetic diversity index analysed. These results do not deny a role for glacial refugia in preserving the spatial pattern of genetic diversity, but emphasize the importance of other factors, like admixture between well-differentiated lineages.
The GLMs (Table 2) indicate that the differences between populations in terms of levels of genetic diversity are best explained by the extent of admixture between distinct lineages occurring within each studied population. A key role for secondary contact and admixture in shaping spatial patterns of population genetic diversity, although not formerly analysed quantitatively in terms of effect size, has emerged in several previous studies for species from the Italian peninsula (see references above), as well as from the other Mediterranean peninsulas of Iberia and the Balkans12,23,24. In most of these studies, however, the secondary contact phase was estimated to have occurred in the late-glacial or post-glacial epochs, or its timeline was not estimated at all. Our Bayesian phylogeographic reconstruction clearly indicated that the range expansion events setting the stage for secondary contacts and gene exchanges initiated, and were most probably completed, well before the last glacial maximum. In the northern Apennine, the co-occurrence of distinct lineages was probably established early in the last interglacial phase, while in the lower Volturno-Calore river basin it most probably occurred before, during the late Middle Pleistocene. This historical scenario has at least one major albeit non-intuitive implication, that is, the spatial patterns of intergradation between lineages and of genetic diversity within populations observed in peninsular Italy is of ancient origin. Therefore, these patterns would have survived at least one glacial-interglacial cycle (probably more) and the substantial range variations that, according to our SDM analyses, affected the B. bufo lineages in peninsular Italy in the Late Pleistocene. As mentioned above, there is an increasing appreciation of the major role and long-term consequences of secondary contact and admixture processes in increasing the genetic diversity of populations, promoting the formation of hotspots, as well as the sharing of adaptive genetic material between divergent lineages. However, in the absence of barriers to gene flow between the interacting lineages, the spatial patterns associated to these historical processes are mostly seen as transient45,46. In fact, the scenario emerging from our results suggests that they might be not so ephemeral as usually thought.
According to the estimated SDMs for the central and southern lineages at the LGM (Fig. 4), a large area of high bioclimatic suitability for the species was located in southern Italy. However, a coastal strip of suitable bioclimatic conditions also occurred along the western side of the peninsula. A comparison of the SDMs estimated for the central and southern lineages suggests that this coastal strip might have run seamless from south to north, preventing a cyclical fragmentation of the B. bufo populations into separate glacial refugia along the south-north axis. Notably, much of this coastal strip is located within areas that are presently below the sea-level, indicating that it acted as a "true refugium" sensu Recuero & García-París47, that is, as an area previously unoccupied by the species, where lineages retreat once their range becomes unsuitable due to paleoclimatic changes.
Together with the very limited and highly fragmented distribution of the inferred areas of stability (see Fig. 4), this scenario might also explain why our correlative models found no evidence of a minimal role for glacial refugia and climatically stable areas in explaining the observed pattern of population genetic diversity. Indeed, we didn’t find evidence of a real “sanctuary-type” refugium, that is, of a large area (or multiple areas) of persistently suitable bioclimatic conditions, where populations persisted over multiple episodes of climatic change47. Instead, throughout most of the peninsula, B. bufo populations likely moved from interior areas (and probably higher altitudes) toward coastal regions and back, in response to paleoclimatic oscillations, through small-scale range migrations that did not erase previously formed patterns of genetic variation. Under this scenario, glacial refugia contributed to preserving levels and patterns of genetic diversity across glacial-interglacial cycles, but not to their formation.
Finally, our results extend previous findings concerning the possible location of glacial refugia within areas that are currently covered by the sea. Indeed, although the occurrence of such areas has already been postulated, they have been mostly linked to insular geographic settings48,49,50, or to continental areas where wide coastal lowlands opened following glacial sea-level drops35,43,51,52. In line with these previous studies, our SDMs for the LGM suggested a possible area of suitable bioclimatic conditions for the southern lineage at the southern edge of the wide coastal lowland now covered by the Adriatic Sea. However, as discussed above, we also found evidence of a narrow glacial refugium along the Tyrrhenian (glacial) coast, which is at the same time an unprecedented but not completely unexpected result. Indeed, paleoenvironmental reconstructions based on palynological, microfossils, and sedimentological data, show the existence of areas of high ecological stability along the western coastal plains of the peninsula53,54, where the effects of climate change were mitigated54. Our results suggest that the importance of these narrow coastal refugia for temperate animal species might have been overlooked in previous phylogeographic studies of co-distributed species35,36,51.
Conclusions
Why are hotpots of genetic diversity so hot? From the perspective of the common toad B. bufo in peninsular Italy the answer is they are because they have been melting pots. Contrary to our expectations, based on previous studies of co-distributed species, we did not find a hotspot of genetic diversity in the southernmost area of the Italian peninsula, a well-documented glacial refugium for a broad range of taxa. Instead, populations from this area were among the least variable. Overall, spatial patterns of population genetic diversity were linked with the extent of admixture between distinct intraspecific lineages rather than with climatically stable areas. This highlights a general principle emerging from the documentation of concordant phylogeographic patterns in species with disparate evolutionary histories in southern European peninsulas: higher levels of genetic diversity mark populations with substantial genetic contributions from multiple differentiated lineages, irrespective of their location regarding glacial refugia.
Materials and methods
Sampling and laboratory procedures
We collected 563 Bufo bufo individuals from 78 sampling localities (Table 1 and Fig. 1). Tissue samples were collected as tail- or toe-clips from tadpoles or adult individuals respectively, which were then released in the respective collection sites. Samples were stored in 95% ethanol until DNA extraction. Field works, collection of tissues, and the experimental protocols were approved by the Italian Ministry of Environment (Permit Numbers: DPN-2009-0026530) and were performed in accordance with the relevant guidelines and regulations (including ethics guidelines and regulations).
DNA extractions were carried out using commercial kits (ZYMO RESEARCH). We amplified by polymerase chain reaction (PCR) two mitochondrial DNA (mtDNA) fragments, a 722-bp fragment of the Cytochrome B gene (CytB) and a 517-bp fragment of the mitochondrial 16 s rRNA gene. Amplifications were performed in a 15-μL reaction volume following protocols described in Recuero et al.26. Purification and sequencing of the PCR products were conducted by Macrogen Inc. (http://www.macrogen.com), using an ABI PRISM 3730 sequencing system (Applied Biosystems). Electropherograms were checked by eye using FinchTV 1.4.0 (Geospiza Inc.). All sequences were deposited in the GenBank database (Supplementary Table S1).
Patterns of genetic variation in the nuclear genome were investigated with nine microsatellite loci (Bspi 3.02, Bspi 3.26, Bspi 4.30, Bspi 3.19, Bspi 4.16, Bspi 3.11, Bspi 4.27, Bspi 4.14 and Bspi 4.29), following previously described protocols55. Other markers known from the literature (see55, and references therein) were excluded from the analysis after trials based on 96 individuals showed inconsistent amplification in > 30% of the individuals analysed. Forward primers were fluorescently labelled and PCR products were electrophoresed by Macrogen Inc. on an ABI 3730xl genetic analyser (Applied Biosystems) with a 400-HD-size standard.
Phylogeographic analyses based on mtDNA
MtDNA sequences were aligned using GeneStudio Pro 2.2.0.0 (GeneStudio Inc., Suwanee, GA). Phylogenetic relationships between mtDNA haplotypes were inferred using the maximum likelihood (ML) method implemented in PhyML3.1056, applying the Neighbour Nearest Interchange method for tree improvement and the best substitution model (TrN93 + G) selected by the Smart Model Selection procedure57 under the Bayesian Information Criterion. The robustness of the topology was assessed via 1000 bootstrap pseudo-replicates. The estimated tree topology was then converted into a haplotype genealogy using Haplotype Viewer58.
The ancestral areas of the main B. bufo genetic lineages and the spatial and temporal patterns of diffusion throughout their range were estimated using the Bayesian phylogeographic (BP) analysis in continuous space implemented in BEAST 1.859. To avoid any potential bias caused by population structure60, we performed separate analyses with the same settings for each main haplogroup identified by the previous phylogenetic analysis. We set the Bayesian skyline as coalescent tree prior61, MCMCs with length of 200 million generations sampling every 20 000 generations, “Cauchy” as spatial diffusion model59,62, a strict molecular clock model, and the fossil-calibrated substitution rate of 5.5 × 10–9 substitutions/year26. Geographical coordinates were provided for each individual, applying a jitter of ± 0.001° to duplicated coordinates. Trace files were inspected using Tracer 1.663 to evaluate the Effective Sample Size (ESS) of the estimated parameters, the appropriate burn-in, and the convergence between runs. Finally, the full posterior sample of trees was analysed in SPREAD 1.0.764, in order to estimate the ancestral area for each lineage (i.e. the geographic location of its most recent common ancestor).
Population structure and genetic diversity based on microsatellite data
Microsatellite data were analysed using GENEMAPPER 4.1. Micro-Checker 2.2.365 was used to test for the presence of null alleles and large-allele dropout. Allelic frequencies, tests for deviations from the expected Hardy–Weinberg and linkage equilibria, and estimates of allelic richness (Ar; computed using the rarefaction method) and the mean observed and expected heterozygosity (Ho and He) were computed using the diveRsity R package66, after excluding populations of sample size n < 4 in at least one locus (localities 12, 13, 14, 20, and 70).
The population genetic structure was evaluated by means of the Bayesian clustering algorithm implemented in TESS 2.3.1, with the geographical origin of individuals as prior information67. The analysis was carried out by modelling admixture using a conditional autoregressive model, and consisted of 100 replicates for each K value (i.e. the number of clusters) between 2 and 10. Each replicate was 50 000 steps long, and the first 20 000 steps were discarded as burn-in. The spatial interaction parameter was kept at the default value (0.6), with the update option activated. The model that best fit the data was selected using the deviance information criterion (DIC). DIC values were averaged over 100 replicates for each value of K, and the best K value was selected as the one at which the average DIC reached a plateau. For the selected K value, the estimated admixture proportions of the 10 runs with the lowest DIC were averaged using CLUMPP 1.1.268.
Identification of glacial refugia
Locations of glacial refugia during the Last Glacial Maximum (LGM) were estimated with species distribution models (SDM) calibrated on the current climate and projected on the LGM. All SDMs were developed using the biomod2 R package, following an ensemble forecasting approach69. To account for intraspecific variability70,71,72, we calibrated distinct SDMs for the central and southern lineages, because they are the only lineages whose geographic distribution has been fully resolved (see Results section). Occurrence data for the entire geographic distribution of each lineage were obtained by pooling field data collected during this study together with data from: Stoch73, Global Biodiversity Information Facility (GBIF.org, 06 April 2017—GBIF Occurrence Download https://doi.org/10.15468/dl.wyiqnj), Observado (www.observation.org). Occurrence data from Stoch73 were validated with our own field observations or by matching toponomy with topography and satellite imagery information, reaching precision ≥ 30 arcseconds (≈1 km at the equator); occurrence data from GBIF were verified by inspecting associated toad pictures. Duplicated records and data georeferenced with uncertainty ≥ 30 arcseconds (≈1 km at the equator) were removed.
To limit spatial autocorrelation among occurrences74, we thinned the raw occurrences using the spThin R package75, obtaining five alternative calibration datasets for each evolutionary lineage. We thinned the occurrences considering the nearest neighbour distance expected in the study area with our sampling effort for a random sample (5.97 km for the central lineage, 9.59 km for the southern lineage along the peninsula, 15.06 km for the southern lineage in Sicily). To limit the effect of sampling bias, we used a target-group approach to inform the selection of background data points76. We collected all data available for reptiles and amphibians available from the same references and for the same study area, obtaining a total of 12,360 unique records; these records were divided following the geographical distribution of each lineage and used in model calibration as background points.
SDMs were calibrated considering the full set of 19 bioclimatic variables (current climate) obtained at 30-arc-second resolution from the WorldClim database (http:\\www.worldclim.org77). We did not include in the analyses elevation data, which are highly collinear with temperature and would represent a problem for model transferability in time74, or NDVI (a satellite based index of the photosyntetically active vegetation), for which no corresponding layer exists back in time. To avoid issues linked to multicollinearity73, we performed a variance inflation factor (VIF) analysis, retaining only variables with VIF < 5. The same variables were also obtained for the LGM climate from the same database (at resolution 2.5′) according to three general circulation models (GCM: CCSM4, MIROC-ESM, MPI-ESMP-P).
For each lineage, we calibrated an ensemble SDM considering multiple initial conditions (the five thinned sets of occurrence points coupled with the set of background points and with the same climate layers) and different model classes (generalised linear model, GLM; generalised additive model, GAM; generalised boosted model, GBM; multivariate adaptive regression spline, MARS; maximum entropy model, MAXENT.Phillips). All models were validated using the occurrences excluded in the thinning; in particular we measured the area under the curve (AUC) of the receiver operating characteristic (ROC) and calculated the probability threshold which maximize the true skill statistics (maxTSS78). The final ensemble model was calculated as the AUC-weighted average of all models with AUC value greater than 0.779 and then projected under the current climate over the study area.
Finally, the same models were projected over the study area also considering LGM climate, obtaining a separate ensemble model for each available GCM. Each model was transformed into a binary model using the maxTSS threshold. We defined as putative glacial refugia all pixels in the Italian peninsula classified as one by at least two GCMs.
Predictors of the spatial patterns of genetic diversity
To explore the relative contribution of genetic admixture between divergent lineages and long-term bioclimatic stability within refugia to current levels of population genetic diversity, we used generalised linear models (GLM) carried out by means of the “glm” function in the lme4 R package. Because we could not estimate the geographic location of glacial refugia in northern Italy (see previous section), subsequent analyses were carried out on the reduced dataset including only populations from the Apennine peninsula and Sicily (localities 24–76).
The extent of admixture within each population (i.e. sampling site) was estimated by calculating the Simpson’s diversity index80, based on the number of genetic clusters (i.e. ancestral lineages) and their proportional contribution to the genetic make-up of each sampled population, as previously estimated by the TESS analysis.
In order to address the contribution of long-term bioclimatic stability to levels of population genetic diversity, we estimated two Euclidean distance measures: (i) distance of each sampled population to the nearest area of glacial refugium, as estimated by the SDM analysis; (ii) distance of each population to the nearest “stability area”, defined as a region of overlap between a glacial refugium and an ancestral area (as previously inferred by the Bayesian phylogeographic analysis). The latter regions were considered as our best estimates for the areas of long-term bioclimatic stability (i.e. stability areas), since all the time windows available (i.e. current, LGM and TMRCA) support the species occurrence in these regions. However, it is worth mentioning that this procedure for the identification of stability areas might overestimate the occurrence of these areas, as in the case of repeated population extinctions and recolonizations in time-windows not covered by previous SDM and Bayesian phylogeographic analyses. Euclidean distances separating each studied population from these areas were computed using the function “near” in ArcGIS 10.1 (ESRI).
Two series of GLMs with Gaussian distribution and identity link function were built, one using allelic richness and one using expected heterozygosity as dependent variables. GLMs were conducted using as predictors: Simpson’s index, the distance from the nearest refugium, the distance from the nearest stability area, as well as their interactions. Model comparisons were carried out by means of the Akaike Information Criterion.
Data availability
DNA sequences used in the present study are deposited in GenBank. Supporting Information files are available in the online version of this article and have been deposited at the Dryad Data Repository (https://doi.org/10.5061/dryad.qz612jmc6).
Change history
20 July 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41598-021-94521-w
References
Alberto, F. J. et al. Potential for evolutionary responses to climate change - evidence from tree populations. Glob. Change Biol. 19, 1645–1661 (2013).
Hoffmann, A. A. & Sgró, C. M. Climate change and evolutionary adaptation. Nature 470, 479–485 (2011).
Ørsted, M., Hoffmann, A. A., Rohde, P. D., Sørensen, P. & Kristensen, T. N. Strong impact of thermal environment on the quantitative genetic basis of a key stress tolerance trait. Heredity 122, 315–325 (2019).
Flight, P. A. Phylogenetic comparative methods strengthen evidence for reduced genetic diversity among endangered tetrapods. Conserv. Biol. 24, 1307–1315 (2010).
Hamabata, T. et al. Endangered island endemic plants have vulnerable genomes. Commun. Biol. 2, 1–10 (2019).
Raffard, A., Santoul, F., Cucherousset, J. & Blanchet, S. The community and ecosystem consequences of intraspecific diversity: a meta-analysis. Biol. Rev. 94, 648–661 (2019).
Whitlock, R. Relationships between adaptive and neutral genetic diversity and ecological structure and functioning: a meta-analysis. J. Ecol. 102, 857–872 (2014).
Koricheva, J. & Hayes, D. The relative importance of plant intraspecific diversity in structuring arthropod communities: a meta-analysis. Funct. Ecol. 32, 1704–1717 (2018).
Kettenring, K. M., Mercer, K. L., Reinhardt Adams, C. & Hines, J. Application of genetic diversity-ecosystem function research to ecological restoration. J. Appl. Ecol. 51, 339–348 (2014).
Hewitt, G. M. The structure of biodiversity—insights from molecular phylogeography. Front. Zool. 1, 4 (2004).
Petit, R. J. et al. Glacial refugia: Hotspots but not melting pots of genetic diversity. Science 300, 1563–1565 (2003).
Hewitt, G. M. Mediterranean Peninsulas—the evolution of hotspots. In Biodiversity Hotspots (eds Zachos, F. E. & Habel, J. C.) 123–148 (Springer, Amsterdam, 2011).
Hampe, A. & Petit, R. J. Conserving biodiversity under climate change: The rear edge matters. Ecol. Lett. 8, 461–467 (2005).
Zampiglia, M. et al. Drilling down hotspots of intraspecific diversity to bring them into on-ground conservation of threatened species. Front. Ecol. Evol. 7, 205 (2019).
Cheng, E., Hodges, K. E., Melo-Ferreira, J., Alves, P. C. & Mills, L. S. Conservation implications of the evolutionary history and genetic diversity hotspots of the snowshoe hare. Mol. Ecol. 23, 2929–2942 (2014).
Brown, S. C., Wigley, T. M. L., Otto-Bliesner, B. L., Rahbek, C. & Fordham, D. A. Persistent Quaternary climate refugia are hospices for biodiversity in the Anthropocene. Nat. Clim. Change 10, 244–248 (2020).
Hewitt, G. M. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol. J. Linn. Soc. 58, 247–276 (1996).
Canestrelli, D., Aloise, G., Cecchetti, S. & Nascetti, G. Birth of a hotspot of intraspecific genetic diversity: notes from the underground. Mol. Ecol. 19, 5432–5451 (2010).
Dufresnes, C. et al. Evolutionary melting pots: a biodiversity hotspot shaped by ring diversifications around the Black Sea in the Eastern tree frog (Hyla orientalis). Mol. Ecol. 25, 4285–4300 (2016).
Canestrelli, D., Bisconti, R., Sacco, F. & Nascetti, G. What triggers the rising of an intraspecific biodiversity hotspot? Hints from the agile frog. Sci. Rep. https://doi.org/10.1038/srep05042 (2014).
Carnaval, A. C., Hickerson, M. J., Haddad, C. F. B., Rodrigues, M. T. & Moritz, C. Stability predicts genetic diversity in the Brazilian Atlantic forest hotspot. Science 323, 785–789 (2009).
Hu, J., Huang, Y., Jiang, J. & Guisan, A. Genetic diversity in frogs linked to past and future climate changes on the roof of the world. J. Anim. Ecol. 88, 953–963 (2019).
Gutiérrez-Rodríguez, J., Barbosa, A. M. & Martínez-Solano, Í. Integrative inference of population history in the Ibero-Maghrebian endemic Pleurodeles waltl (Salamandridae). Mol. Phylogenet. Evol. 112, 122–137 (2017).
Gutiérrez-Rodríguez, J., Barbosa, A. M. & Martínez-Solano, Í. Present and past climatic effects on the current distribution and genetic diversity of the Iberian Spadefoot toad (Pelobates cultripes): an integrative approach. J. Biogeogr. 44, 245–258 (2017).
Garcia-Porta, J. et al. Molecular phylogenetics and historical biogeography of the west-palearctic common toads (Bufo bufo species complex). Mol. Phylogenet. Evol. 63, 113–130 (2012).
Recuero, E. et al. Multilocus species tree analyses resolve the radiation of the widespread Bufo bufo species group (Anura, Bufonidae). Mol. Phylogenet. Evol. 62, 71–86 (2012).
Arntzen, J. W., de Vries, W., Canestrelli, D. & Martínez-Solano, I. Hybrid zone formation and contrasting outcomes of secondary contact over transects in common toads. Mol. Ecol. 26, 5663–5675 (2017).
Arntzen, J. W., de Vries, W., Canestrelli, D. & Martínez-Solano, I. Genetic and morphological differentiation of common toads in the Alps and the Apennines. In Evolutionary Biology: A Transdisciplinary Approach (ed. Pontarotti, P.) 1–14 (Springer, Switzerland, 2020).
Lanza, B., Andreone, F., Bologna, M. A., Corti, C. & Razzetti, E. Fauna d’Italia–Amphibia XLII (Calderini, Bologna, 2007).
Barbanera, F. et al. Molecular phylogeography of the asp viper Vipera aspis (Linnaeus, 1758) in Italy: evidence for introgressive hybridization and mitochondrial DNA capture. Mol. Phylogenet. Evol. 52, 103–114 (2009).
Bisconti, R., Porretta, D., Arduino, P., Nascetti, G. & Canestrelli, D. Hybridization and extensive mitochondrial introgression among fire salamanders in peninsular Italy. Sci. Rep. https://doi.org/10.1038/s41598-018-31535-x (2018).
Canestrelli, D., Zangari, F. & Nascetti, G. Genetic evidence for two distinct species within the Italian endemic Salamandrina terdigitata Bonnaterre, 1789 (Amphibia: Urodela: Salamandridae). Herpetol. J. 16, 221–227 (2006).
Salvi, D., Pinho, C. & Harris, D. J. Digging up the roots of an insular hotspot of genetic diversity: Decoupled mito-nuclear histories in the evolution of the Corsican-Sardinian endemic lizard Podarcis tiliguerta. BMC Evol. Biol. https://doi.org/10.1186/s12862-017-0899-x (2017).
Schultze, N. et al. Mitochondrial ghost lineages blur phylogeography and taxonomy of Natrix helvetica and N. natrix in Italy and Corsica. Zool. Scr. 49, 395–411 (2020).
Canestrelli, D., Salvi, D., Maura, M., Bologna, M. A. & Nascetti, G. One species, three Pleistocene evolutionary histories: phylogeography of the Italian crested newt Triturus carnifex. PLoS ONE https://doi.org/10.1371/journal.pone.0041754 (2012).
Canestrelli, D., Cimmaruta, R. & Nascetti, G. Phylogeography and historical demography of the Italian treefrog, Hyla intermedia, reveals multiple refugia, population expansions and secondary contacts within peninsular Italy. Mol. Ecol. 16, 4808–4821 (2007).
Canestrelli, D., Sacco, F. & Nascetti, G. On glacial refugia, genetic diversity, and microevolutionary processes: Deep phylogeographical structure in the endemic newt Lissotriton italicus. Biol. J. Linn. Soc. 105, 42–55 (2012).
Canestrelli, D., Cimmaruta, R., Costantini, V. & Nascetti, G. Genetic diversity and phylogeography of the Apennine yellow-bellied toad Bombina pachypus, with implications for conservation. Mol. Ecol. 15, 3741–3754 (2006).
Canestrelli, D., Cimmaruta, R. & Nascetti, G. Population genetic structure and diversity of the Apennine endemic stream frog, Rana italica - Insights on the Pleistocene evolutionary history of the Italian peninsular biota. Mol. Ecol. 17, 3856–3872 (2008).
Chiocchio, A. et al. Population genetic structure of the bank vole Myodes glareolus within its glacial refugium in peninsular Italy. J. Zool. Syst. Evol. Res. 57, 959–969 (2019).
Podnar, M., Mayer, W. & Tvrtković, N. Phylogeography of the Italian wall lizard, Podarcis sicula, as revealed by mitochondrial DNA sequences. Mol. Ecol. 14, 575–588 (2005).
Vai, F. & Martini, I. P. Anatomy of an Orogen: The Apennines and Adjacent Mediterranean Basins (Springer, Berlin, 2013).
Canestrelli, D., Verardi, A. & Nascetti, G. Genetic differentiation and history of populations of the Italian treefrog Hyla intermedia: Lack of concordance between mitochondrial and nuclear markers. Genetica 130, 241–255 (2007).
Stefani, F., Galli, P., Crosa, G., Zaccara, S. & Calamari, D. Alpine and Apennine barriers determining the differentiation of the rudd (Scardinius erythrophthalmus L.) in the Italian peninsula. E. Freshw. Fish 13, 168–175 (2004).
Dynesius, M. & Jansson, R. Persistence of within-species lineages: a neglected control of speciation rates. Evolution 68, 923–934 (2014).
Endler, J. A. Geographic Variation, Speciation, and Clines (Princeton University Press, Princeton, 1977).
Recuero, E. & García-París, M. Evolutionary history of Lissotriton helveticus: Multilocus assessment of ancestral vs. recent colonization of the Iberian Peninsula. Mol. Phylogenet. Evol. 60, 170–182 (2011).
Bisconti, R., Canestrelli, D., Colangelo, P. & Nascetti, G. Multiple lines of evidence for demographic and range expansion of a temperate species (Hyla sarda) during the last glaciation. Mol. Ecol. 20, 5313–5327 (2011).
Salvi, D., Schembri, P. J., Sciberras, A. & Harris, D. J. Evolutionary history of the Maltese wall lizard Podarcis filfolensis: Insights on the “Expansion-Contraction” model of Pleistocene biogeography. Mol. Ecol. 23, 1167–1187 (2014).
Senczuk, G. et al. Evolutionary and demographic correlates of Pleistocene coastline changes in the Sicilian wall lizard Podarcis wagleriana. J. Biogeogr. 46, 224–237 (2019).
Canestrelli, D. & Nascetti, G. Phylogeography of the pool frog Rana (Pelophylax) lessonae in the Italian peninsula and Sicily: Multiple refugia, glacial expansions and nuclear-mitochondrial discordance. J. Biogeogr. 35, 1923–1936 (2008).
Porretta, D. et al. Southern crossroads of the Western Palaearctic during the Late Pleistocene and their imprints on current patterns of genetic diversity: Insights from the mosquito Aedes caspius. J. Biogeogr. 38, 20–30 (2011).
Amorosi, A. et al. The Holocene evolution of the Volturno River coastal plain (southern Italy). J. Mediterr. Earth Sci. 7, 11 (2013).
Lucchi, M. R. Vegetation dynamics during the Last Interglacial-Glacial cycle in the Arno coastal plain (Tuscany, western Italy): location of a new tree refuge. Quat. Sci. Rev. 27, 2456–2466 (2008).
Trujillo, T., Gutiérrez-Rodríguez, J., Arntzen, J. W. & Martínez-Solano, I. Morphological and molecular data to describe a hybrid population of the Common toad (Bufo bufo) and the Spined toad (Bufo spinosus) in western France. Contrib. Zool. 8, 1–10 (2017).
Guindon, S. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010).
Lefort, V., Longueville, J.-E. & Gascuel, O. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 34, 2422–2424 (2017).
Salzburger, W., Ewing, G. B. & Von Haeseler, A. The performance of phylogenetic algorithms in estimating haplotype genealogies with migration. Mol. Ecol. 20, 1952–1963 (2011).
Lemey, P., Rambaut, A., Welch, J. J. & Suchard, M. A. Phylogeography takes a relaxed random walk in continuous space and time. Mol. Biol. Evol. 27, 1877–1885 (2010).
Heller, R., Chikhi, L. & Siegismund, H. R. The confounding effect of population structure on bayesian skyline plot inferences of demographic history. PLoS ONE 8(5), e62992. https://doi.org/10.1371/journal.pone.0062992 (2013).
Drummond, A. J., Rambaut, A., Shapiro, B. & Pybus, O. G. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 22, 1185–1192 (2005).
Paradis, E., Baillie, S. R. & Sutherland, W. J. Modeling large-scale dispersal distances. Ecol. Model. 151, 279–292 (2002).
Rambaut, A., Suchard, M. A., Xie, D. & Drummond, A. J. (2014). Tracer v1.6. Available at: http://beast.bio.ed.ac.uk/Tracer.
Bielejec, F., Rambaut, A., Suchard, M. A. & Lemey, P. SPREAD: spatial phylogenetic reconstruction of evolutionary dynamics. Bioinformatics 27, 2910–2912 (2011).
Van Oosterhout, C., Hutchinson, W. F., Wills, D. P. & Shipley, P. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes. 4, 535–538 (2004).
Keenan, K., Mcginnity, P., Cross, T. F., Crozier, W. W. & Prodöhl, P. A. DiveRsity: An R package for the estimation and exploration of population genetics parameters and their associated errors. Methods. Ecol. Evol. 4, 782–788 (2013).
Chen, C., Durand, E., Forbes, F. & François, O. Bayesian clustering algorithms ascertaining spatial population structure: a new computer program and a comparison study. Mol. Ecol. Notes 7, 747–756 (2007).
Jakobsson, M. & Rosenberg, N. A. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23, 1801–1806 (2007).
Araújo, M. B. & New, M. Ensemble forecasting of species distributions. Trends Ecol. Evol. 22, 42–47 (2007).
Lecocq, T., Harpke, A., Rasmont, P. & Schweiger, O. Integrating intraspecific differentiation in species distribution models: consequences on projections of current and future climatically suitable areas of species. Divers. Distrib. 25, 1088–1100 (2019).
Hällfors, M. H. et al. Addressing potential local adaptation in species distribution models: implications for conservation under climate change. Ecol. Appl. 26, 1154–1169 (2016).
Pearman, P. B., D’Amen, M., Graham, C. H., Thuiller, W. & Zimmermann, N. E. Within-taxon niche structure: niche conservatism, divergence and predicted effects of climate change. Ecography 33, 990–1003 (2010).
Stoch, F. CKmap 5.3.8, http://www.faunaitalia.it/documents/CKmap_54.zip. Ministero dell’Ambiente e della Tutela del Territorio, Direzione Protezione della Natura (2000–2005)
Araújo, M. B. et al. Standards for distribution models in biodiversity assessments. Sci. Adv. 5(1), eaat4858. https://doi.org/10.1126/sciadv.aat4858 (2019).
Aiello-Lammens, M. E., Boria, R. A., Radosavljevic, A., Vilela, B. & Anderson, R. P. spThin: An R package for spatial thinning of species occurrence records for use in ecological niche models. Ecography 38, 541–545 (2015).
Ranc, N. et al. Performance tradeoffs in target-group bias correction for species distribution models. Ecography 40(9), 1076–1087 (2017).
Hijmans, R. J., Cameron, S. E., Parra, J. L., Jones, P. G. & Jarvis, A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 25, 1965–1978 (2005).
Freeman, E. A. & Moisen, G. G. A comparison of the performance of threshold criteria for binary classification in terms of predicted prevalence and kappa. Ecol. Model. 217, 48–58 (2008).
Swets, J. A. Measuring the accuracy of diagnostic systems. Science 240, 1285–1293 (1988).
Simpson, E. H. Measurement of diversity. Nature 163, 688 (1949).
Acknowledgements
We are grateful to Mauro Zampiglia, Francesco Paolo Caputo, Gaetano Aloise, and Francesco Pellegrino for helping in sampling activities. This research was supported by grants from the Italian Ministry of Education, University and Research (PRIN Project 2012FRHYRA).
Author information
Authors and Affiliations
Contributions
D.C., J.W.A. and I.M.S. conceived the study. A.C., D.C. and W.d.V. performed sample collection. A.C. performed laboratory experiments. A.C., D.C. and R.B. analysed the genetic data. A.P. and L.M. performed SDM. A.C. and D.C. wrote the paper, with inputs from all authors. All authors reviewed and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this Article was revised: The original version of this Article contained an error in Figure 1 and Figure 3 where parts of the figures were incorrectly captured.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chiocchio, A., Arntzen, J.W., Martínez-Solano, I. et al. Reconstructing hotspots of genetic diversity from glacial refugia and subsequent dispersal in Italian common toads (Bufo bufo). Sci Rep 11, 260 (2021). https://doi.org/10.1038/s41598-020-79046-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-79046-y
- Springer Nature Limited
This article is cited by
-
Chloroplast DNA phylogeography reveals genetic divergence of bermudagrass along latitudinal and longitudinal gradients in China
Genetic Resources and Crop Evolution (2024)
-
Testing assertions of widespread introgressive hybridization in a clade of neotropical toads with low mate selectivity (Rhinella granulosa species group)
Heredity (2023)
-
Unveiling a hotspot of genetic diversity in southern Italy for the endangered Hermann’s tortoise Testudo hermanni
BMC Ecology and Evolution (2022)
-
Across the Gobi Desert: impact of landscape features on the biogeography and phylogeographically-structured release calls of the Mongolian Toad, Strauchbufo raddei in East Asia
Evolutionary Ecology (2022)
-
Verification of hotspots of genetic diversity in Korean population of Grateloupia asiatica and G. jejuensis (Rhodophyta) show low genetic diversity and similar geographic distribution
Genes & Genomics (2021)