Abstract
HPV (Human papilloma virus) is a kind of small double-stranded DNA viruses which is extremely associated with different cancers. The roles HPV plays in the host were gradually identified through the interaction between it (including its early genes) and host RNA. In recent years, increasing numbers of studies in HPV-related cancers have been published showing the relationship between HPV and host RNA. Here, we present a database named HRRD, which contains the regulatory relationship between HPV and RNA (mRNA, miRNA and lncRNA). The information was extracted from 10,761 papers in PubMed (up to December 1st, 2019). In addition, the sequence map of HPV (198 genotypes) is also contained. HRRD was designed as a user-friendly web-based interface for data retrieval. It integrated the information of interaction between HPV and RNA, which reflects the relationship between HPV and host. We hope HRRD will further provide a comprehensive understanding of HPV in carcinogenesis and prognosis. HRRD is freely accessible at www.hmuhrrd.com/HRRD.
Similar content being viewed by others
Introduction
Human papilloma viruses (HPVs) are a group of double-stranded circular DNA viruses with about 8000 nucleotides in size. More than 200 HPV types have been characterized to date, including some high-risk types (16, 18, 31, 33 and so on). HPV genome is composed of six early gene (E1, E2, E4, E5, E6 and E7) open reading frames (ORFs), two late gene (L1 and L2) ORFs and a non-coding long control region (LCR). Distinct genes perform different functions1. After HPVs infect the basal cells of epithelia, they employ cellular machinery to replicate and survive in host cells2. With the persistent infection, HPV and the released viral genome will lead to squamous intraepithelial lesions, even cancers3, such as cervical cancer4 and head and neck cancer5. HPV not only plays an important role in carcinogenesis, but also participate in host tumor immunity, prognosis, drug resistance and so on. All these functions are achieved through the interaction between HPV and host RNA.
Host RNA can be divided into coding RNA and non-coding RNA. mRNA is the main type of coding RNA, while miRNA and lncRNA are two kinds of non-coding RNA which are popular in the field of research. HPVs influence host cells (normal/cancer cells) by regulating the expression of these RNAs (Fig. 1). In normal cells, HPV mainly affects cell cycle, apoptosis and differentiation. For example, E6 promotes p53 degradation and inhibits cell apoptosis, while E7 binds to pRb and promotes cell growth and differentiation6. HPV activates miR-34 to participate in the cell apoptosis7. In addition, the expression of HOTAIR was significantly down-regulated by E7 to cause cervical cancer8. In cancer cells, HPV participates in cancer metastasis, prognosis, immune response and so on. For instance, HPV activates TGF-β and IL-10 expression to activate the host immunity9. HPV promotes tumor metastasis by affecting various miRNAs in HPV-related cancers10. Besides, there are many other RNA changes caused by HPV and its oncogenes in HPV-related cancers. By affecting the expression of host RNA, HPV can lead to the occurrence and development of cancers, and has a huge impact on the treatment, immunity and prognosis of cancers. Moreover, it has also been reported recently that host RNA can affect the expression of HPV in converse.
In the recent past, some databases about HPV have been published. PaVE11 is a database about papilloma virus (including HPV). HPVdb12 is a Human Papillomavirus T cell Antigen Database. HPVbase13 shows the integration information of HPV. However, these databases only focus on the biological characteristics of HPV itself, such as its type and sequence information, but do not collect any information about the HPV-infected host. Despite of numerous experimental studies in the field, there are no computational resources with a unique focus on the relationship between HPV and host RNA. Hence, it is important and necessary to establish a proper multi-comparative platform to show this mutual relationship. Here, we present HRRD, a database of the relationship between HPV and host RNA, which contains the existing studies on HPV and host RNA. It will contribute to providing a comprehensive web-based resource for HPV and host, and data support for HPV-related research.
Methods
Implementation
HRRD was developed with Eclipse (https://www.eclipse.org), one of the open-source integrated development environment (IDE). The database manage system was MySQL (https://www.mysql.com) which stores the relationship data model. Java Server Pages (JSP) in the web tier was used to present dynamic responses to clients. The client tier was made up of static and dynamic html pages including forms, tables, charts, and embedded JavaScripts. The model-view-controller (MVC) model web application was managed by Struts (https://struts.apache.org). All plots and web crawler were generated with python (version 3.8.1). HRRD is implemented and hosted on a Windows operating system, with Tomcat and MySQL (version 5.6.19), for serving static and dynamic content and providing the features for data management, respectively (Fig. 2).
Flow chart of database construction. Files of HPV sequence were downloaded from Genbank. Related literatures searched by keywords were downloaded via web crawler in Pubmed. HPV-RNA interaction information was extracted manually. All the information was stored in HRRD, then computed or analyzed using scripts with outputs visualized in figures, txt and tables.
Data collection and processing
HPV-related search keywords include: HPV, human papillomavirus and human papilloma virus. RNA-related search keywords include: RNA, miRNA (microRNA), mRNA, lncRNA (long non-coding RNA), gene. Articles with the combination of keywords were searched on PubMed (excluding reviews and epidemiological studies). Web crawler got the research literatures with the keywords above. After deleting the epidemiological survey, review and the duplications, there were 10,761 articles remaining (up to December 1st, 2019). The whole genome information of HPV (198 genotypes with specific sequence information) was downloaded from NCBI in genbank format.
The information of the relationship between HPV and host RNA was extracted from articles, containing the PMID number, cancer type, HPV genotype, HPV oncogene, RNA type and name, method, material and the regulatory relationship between HPV and host RNA (action direction & action type). The information is obtained by reading the literature manually. The collected data were checked and unified, and finally summarized into Excel form. There are totally 784 pieces of information extracted from the articles, including 13 pieces of lncRNA, 195 pieces of miRNA, and others are mRNA.
The sequence information of different HPV was extracted from the genbank files, including the full-length genome of HPV and the location of its oncogenes. The information of HPV sequence was stored in the database in TXT format.
Results
Relationship page
The principal function of HRRD is query. The information about the relationship between HPV and host RNA is stored in the database. In the Relationship page, there are three input boxes with examples for users to search. Users can search by one to three conditions like HPV genotype, HPV gene or RNA name to get a list containing the relevant content of users searched. The list would contain following information: HPV genotype, HPV gene, RNA type and name, action direction and type, cancer type, PMID, method and material. Users can jump to the article summary in PubMed by clicking the PMID number. “HPV to host” means HPV affects host RNA expression, while “host to HPV” means host RNA influences HPV expression. “Activation” refers to up-regulated expression, while “Suppression” means down-regulated expression. The method shows whether this message was concluded from experimental verification or data analysis (Fig. 3C). The result was sorted by the name of RNA. The export button can help users to export the searched results in XLS format.
Sequence page
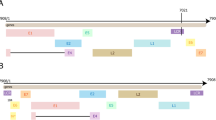
The genome information of HPVs is stored in the database. In the Sequence page, the table shows all species of HPV stored in HRRD. Users can input different genotype of HPV in the search box and click the search button to search HPV in the table below. Users can go to the result page by the plot button or clicking the HPV ID in the table (Fig. 3B). The result page is divided into two parts. The upper part displays the linear viewer of the corresponding HPV sequence, and the complete sequence of the HPV is shown in the lower part (Fig. 3D). The picture and sequence of HPV genome can be accessed directly.
Other pages
Other pages contain some other ancillary functions. A simple Home page was designed (Fig. 3A) with a brief introduction of HRRD. The scrolling picture on the Home page shows HPV, the process of HPV infection and the effect of HPV on host cells. Some frequently asked questions and answers about HRRD are given in the FAQ page. In Links page, there shows the hyperlinks and brief introduction of some HPV-related databases like PaVE, HPVdb, HPVbase and NCBI. If necessary, users can visit the corresponding website. In the Download page, there is a software hyperlink named DisV-HPV16 (version I), which was developed for detecting HPV16 and its oncogenes expression in RNA-sequencing data.
Discussion and future development
To our knowledge, HRRD is the first database designed for the relationship between HPV and RNA specifically. To ensure the accuracy of data, the information was manually extracted from relevant literature. HRRD describes the sequence map of HPV and incorporates some links to HPV-related databases so that users can search and get relevant information quickly and easily. We chose RNA as our research object, because RNA is unstable and susceptible to changes and mutates by viruses, and it is the first step in protein synthesis. For example, if researchers detect HPV infection in cancer, HRRD can help to find out whether HPV affects the key RNA and causes the carcinogenesis. In addition, if researchers want to find some key RNA in HPV-related cancer, HRRD can help to do preliminary screening and query. With the plenty resources, it will provide candidate biomarkers for HPV-related carcinogenesis and prognosis and identify potential targets for molecular therapy. We anticipate it will facilitate an insight into HPV and host.
Despite the database has contained data up to December 1st, 2019, a rapidly growing number of relevant researches are emerging from then. HRRD will be updated regularly by scanning newly published literatures. To ensure the timeliness of data, we plan to use data mining and machine learning to obtain information from the literatures. At the same time, we will update the software with modified function and expanded the scope of application. These updates will be implemented in the near future.
Data availability
HRRD is freely accessible at www.hmuhrrd.com/HRRD.
References
Tsakogiannis, D., Gartzonika, C., Levidiotou-Stefanou, S. & Markoulatos, P. Molecular approaches for HPV genotyping and HPV-DNA physical status. Expert Rev. Mol. Med. 19, e1. https://doi.org/10.1017/erm.2017.2 (2017).
Hong, S. & Laimins, L. A. Regulation of the life cycle of HPVs by differentiation and the DNA damage response. Future Microbiol. 8, 1547–1557. https://doi.org/10.2217/fmb.13.127 (2013).
Brianti, P., De Flammineis, E. & Mercuri, S. R. Review of HPV-related diseases and cancers. New Microbiol. 40, 80–85 (2017).
Doorbar, J. Molecular biology of human papillomavirus infection and cervical cancer. Clin. Sci. (Lond.) 110, 525–541. https://doi.org/10.1042/CS20050369 (2006).
Rautava, J. & Syrjanen, S. Biology of human papillomavirus infections in head and neck carcinogenesis. Head Neck Pathol. 6(Suppl 1), S3–S15. https://doi.org/10.1007/s12105-012-0367-2 (2012).
Maxwell, J. H., Grandis, J. R. & Ferris, R. L. HPV-associated head and neck cancer: Unique features of epidemiology and clinical management. Annu. Rev. Med. 67, 91–101. https://doi.org/10.1146/annurev-med-051914-021907 (2016).
Raver-Shapira, N. et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol. Cell 26, 731–743. https://doi.org/10.1016/j.molcel.2007.05.017 (2007).
Sharma, S. et al. Bridging links between long noncoding RNA HOTAIR and HPV oncoprotein E7 in cervical cancer pathogenesis. Sci. Rep. 5, 11724. https://doi.org/10.1038/srep11724 (2015).
Barros, M. R. Jr. et al. Activities of stromal and immune cells in HPV-related cancers. J. Exp. Clin. Cancer Res. 37, 137. https://doi.org/10.1186/s13046-018-0802-7 (2018).
Santos, J. M. O., da Silva, S. P., Costa, N. R., da Costa, R. M. G. & Medeiros, R. The role of microRNAs in the metastatic process of high-risk HPV-induced cancers. Cancers (Basel) https://doi.org/10.3390/cancers10120493 (2018).
Van Doorslaer, K. et al. The Papillomavirus episteme: A central resource for papillomavirus sequence data and analysis. Nucleic Acids Res. 41, D571–D578. https://doi.org/10.1093/nar/gks984 (2013).
Zhang, G. L. et al. HPVdb: A data mining system for knowledge discovery in human papillomavirus with applications in T cell immunology and vaccinology. Database 2014, 31. https://doi.org/10.1093/database/bau031 (2014).
Kumar Gupta, A. & Kumar, M. HPVbase—A knowledgebase of viral integrations, methylation patterns and microRNAs aberrant expression: As potential biomarkers for Human papillomaviruses mediated carcinomas. Sci. Rep. 5, 12522. https://doi.org/10.1038/srep12522 (2015).
Acknowledgements
This study was supported by the Natural Science Foundation of China (81501737), Outstanding Youth Foundation of Heilongjiang Province (jc2018023) and Student innovation fund of Harbin Medical University (YJSKCX2018-79HYD). We thank all students helping us in Harbin Medical University: Yinnan Zhang, Lechun Ou, Xinyi Xie and Congcong Zhang. We gratefully thank Wu Lien-Ten Institute for providing the computational server used in code debugging.
Author information
Authors and Affiliations
Contributions
L.L.W. conceived the study and supervised the project. H.S., X.Y.D. and F.M. read literature. B.Z.W. and T.Y.L. checked data. B.Q.Y. and S.W.Z. designed the system architecture. BQY wrote the source code. B.Q.Y. drafted the manuscript. S.Y.Y. and T.Y.L. revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yan, B., Zhang, S., Yu, S. et al. HRRD: a manually-curated database about the regulatory relationship between HPV and host RNA. Sci Rep 10, 19586 (2020). https://doi.org/10.1038/s41598-020-76719-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-76719-6
- Springer Nature Limited