Abstract
Activation of toll-like receptors (TLR1, TLR5, TLR6) and downstream markers (CCR1, MAPK14, ICAM1) leads to increased systemic inflammation. Our objective was to study the association between the gene expression levels of these six genes and lung function (Forced Expiratory Volume in one second (FEV1), Forced Vital Capacity (FVC) and FEV1/FVC). We studied gene expression levels and lung function in the Coronary Artery Risk Development in Young Adults study. Spirometry testing was used to measure lung function and gene expression levels were measured using the Nanostring platform. Multivariate linear regression models were used to study the association between lung function measured at year 30, 10-year decline from year 20 to year 30, and gene expression levels (highest quartile divided into two levels – 75th to 95th and>95th to 100th percentile) adjusting for center, smoking and BMI, measured at year 25. Year 30 FEV1 and FVC were lower in the highest level of TLR5 compared to the lowest quartile with difference of 4.00% (p for trend: 0.04) and 3.90% (p for trend: 0.05), respectively. The 10-year decline of FEV1 was faster in the highest level of CCR1 as compared to the lowest quartile with a difference of 1.69% (p for trend: 0.01). There was no association between gene expression and FEV1/FVC. Higher gene expression levels in TLR5 and CCR1 are associated with lower lung function and faster decline in FEV1 over 10 years, in a threshold manner, providing new insights into the role of inflammation in lung function.
Similar content being viewed by others
Introduction
Biomarkers of systemic inflammation such as C-reactive protein (CRP) and fibrinogen have been associated with lower FEV1 and FVC, chronic obstructive pulmonary disease (COPD) and asthma1,2,3,4,5,6. However, these are non-specific inflammatory biomarkers that do not help elucidate the underlying biological pathways that increase systemic inflammation. For example, CRP can be modulated by numerous biological pathways such as NF-κB or the JAK/STAT pathway7,8 and activation of any one of these pathways would increase CRP levels. Thus, measuring CRP alone does not identify specific causes of inflammation. A better understanding of specific biological pathways that modulate down-stream inflammatory markers such as CRP may be useful in understanding the pathogenesis of pulmonary diseases such as COPD and asthma. Technological advances have enabled measurement of gene expression levels for thousands of targets simultaneously in epidemiological studies9,10 and can provide novel insights into biological pathways that influence the pathogenesis of chronic diseases.
In this study, we evaluated the pathogen recognition pathway, which is a specific inflammatory pathway, mediated by Toll-Like Receptors (TLRs), which leads to the activation and nuclear translocation of NF-κB that promotes inflammation. TLRs are activated after they specifically recognize Pathogen Associated Molecular Patterns (PAMPs) from a variety of pathogens11,12 and are involved in the pathogenesis of inflammatory diseases such as asthma and COPD13,14. TLR1 and TLR6 are cell surface receptors found on the surface of innate immune cells such as monocytes, myeloid dendritic cells and plasmacytoid dendritic cells, which form heterodimers with TLR2 after activation by bacterial lipopeptides from gram positive bacteria15,16. TLR1/TLR2 heterodimer regulates the expression of CCR1, a chemokine receptor, which upon activation by its ligands such as CCL3, CCL5, CCL7 and CCL23 helps in the recruitment of immune cells to the site of inflammation17,18. TLR1/TLR2 heterodimer also activates ICAM1, which was previously shown to be associated with lung function19,20. TLR5 is another cell surface toll-like receptor which is activated by flagellin, a principal component of bacterial flagellum. TLR5 is present on the surface of monocytes and myeloid dendritic cells21,22 and is involved in the priming of allergic responses to dust allergens promoting asthma23. TLR5, TLR6 and TLR2 activation causes downstream activation of MAPK14 and the NF-κB pathway24,25 that ultimately results in the release of proinflammatory cytokines such as IL-1β, TNF-α and IL-6 and, proliferation of immune cells26.
The primary objective of this study was to evaluate associations between gene expression levels, in mixed white cells in a community-based sample, of six inflammatory markers and lung function defined by FEV1, FVC and FEV1/FVC ratio in the Coronary Artery Risk Development in Young Adults (CARDIA) study. We hypothesized that higher gene expression level of biomarkers associated with increased inflammation would be associated with a lower lung function measurement, and with a faster decline in lung function.
Methods
Study population
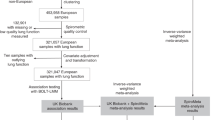
CARDIA is a cohort study with 5115 participants who were recruited at baseline examination during the year 1985-1986 at 4 field centers (Birmingham,AL: Chicago, IL; Minneapolis, MN; and Oakland, CA). The study included approximately equal number of blacks and white; men and women. The follow-up in 2005–2006 (year 20 exam) included 3547 participants (72% of survivors); in 2010–2011 (year 25 exam) was 3499 (72% of survivors) and in 2015–2016 (year 30 exam) was 3358 (71% of survivors). The detailed methods, instruments and quality control procedures for the CARDIA study have been previously described27,28. Demographics, lifestyle habits, physical activity were self-reported using questionnaires. All study methods were carried out in accordance with relevant guidelines and regulations. The CARDIA study is reviewed annually and approved by the internal review boards at Kaiser Permanente Division of Research, Northwestern University, University of Minnesota and University of Alabama at Birmingham. All CARDIA participants provided a signed informed consent before study participation and sign a new informed consent form at every examination.
Three thousand five hundred and forty participants attended the year 20 examination, 3499 participants attended the year 25 examination and 3358 participants attended the year 30 examination. For the cross-sectional analyses using year 25 gene expression levels and year 30 lung function measurements, 2527 participants were included in the analyses after excluding pregnant women (n = 10), participants with missing year 30 lung function data (n = 185), year 25 gene expression measurements (n = 425) and other covariates at year 25 (n = 9 for BMI and n = 54 for smoking). To evaluate association of 10-year decline of lung function from year 20 to year 30 with year 25 gene expression measurements, 2271 participating were included in the analyses after excluding an additional 117 participants with missing lung function data at year 20 in addition to the participants excluded in the cross-sectional analyses. The participants with lung function data were more likely to have lower BMI (29.92 vs. 30.64; p-value = 0.005), lower alcohol consumption (10.91 mL per day vs. 13.22 mL per day; p-value = 0.01), lower C-reactive protein (2.96 vs. 3.84; p-value = 0.0007), had lower percentage of blacks (42.89% vs. 58.49%; p-value <0.0001), had higher percentage of women (58.43% vs. 51.35%; p-value <0.0001) and lower current smokers (12.86% vs. 25.30%; p-value <0.0001) as compared to CARDIA participants who were not included in the analyses (n = 2844). For the sensitivity analysis, we excluded 55 participants with COPD and 476 participants with asthma in years 25 and 30 when evaluating the cross-sectional association between year 30 lung function and year 25 gene expression. We also excluded 47 participants with COPD and 442 participants with asthma at years 20, 25 and 30 to evaluate the longitudinal association between 10-year decline of lung function and year 25 gene expression.
Spirometry
Spirometry was performed using a dry rolling-seal OMI spirometer (Viasys Corp, Loma Linda, CA) at year 20 examination and a portable spirometer EasyOne Diagnostic, NDD Medical Technologies, Andover,MA) at year 30 following the American Thoracic Society Guidelines29. Daily checks for leaks, volume calibration with a 3-liter syringe and weekly calibration in the 4–7 litre range were undertaken to minimize methodological artifacts between exams. We analyzed FVC and FEV1 as the maximum of five satisfactory maneuvers and represented as percent of predicted30. In almost all cases, the maximum and second highest maneuvers agreed to within 150 ml.
Gene expression analysis
Whole blood was collected in the PAXgene Blood RNA tubes (Qiagen Inc., Germantown, MD) at the year 25 examination. mRNA was isolated using the PAXgene Blood RNA kit (Qiagen Inc., Germantown, MD) at the Molecular Epidemiology and Biomarker Research Laboratory (MEBRL) according to the manufacturer’s instructions.
The nCounter analysis system (Nanostring Inc., Seattle, WA) was used to measure gene expression levels using mRNA obtained from whole blood collected in a PAXgene tube. The nCounter system utilized two unique~50 base probes, or “code set” per mRNA, a Capture probe that immobilized the probe/target complex to the nCounter cartridge and a Reporter probe for signal detection. The digitally captured color codes were counted and tabulated for each target molecule. As a quality control measure internal control (positive and negative) code sets were used. The positive control probes are mixed in a varying concentration corresponding to expression levels of most mRNAs of interest, and the negative control probes are used to estimate the non-specific background in the experiment.
Normalization of the gene expression was done with a combination of positive control normalization and CodeSet Content Normalization using housekeeping genes to correct major sources of error including pipetting errors, instrument scan resolution, batch variations and sample input variability. The positive control normalization factor was the ratio of the average ERCC positive control intensity seen across all samples divided by the positive control within an individual sample. The geometric mean of the 5 housekeeping genes (B2M, GAPDH, GNB1, HPRT1, PGK1) was calculated for each sample and the average geometric mean across all samples divided by the geometric mean of the housekeeping genes for each sample was used to normalize gene expression levels. In addition, the average count for each target is calculated using all the samples probed within a particular CodeSet and this average is used to calculate a CodeSet normalization factor to account for the variation in the efficiency at capturing and scanning each unique target type. The raw counts of the gene expression of sample were first multiplied by the sample specific positive control normalization factor, then by the housekeeping gene normalization factor and the CodeSet normalization factor to obtain the final gene expression counts.
Measurement of covariates and determination of asthma/COPD status
Smoking status was measured using questionnaires. Never smokers were defined as participants who reported never having used any tobacco products such as cigarettes, cigars or not having smoked regularly for at least three months. Former smokers were defined as having smoked regularly for at least three months but are not currently smoking regularly. Current smokers were defined as having smoked regularly for at least three months and are smoking regularly currently. BMI was calculated using the height and weight variables as weight (in kg) divided by height (in meters) squared. BMI and smoking status measured at year 25 were used in the analysis. Asthma was defined by self-report of whether the participants were ever diagnosed as having asthma or if they were currently taking any asthma medications. COPD was also self-reported by the participants.
Statistical methods
Selected year 25 characteristics among 5 levels of TLR5 gene expression were compared using chi-square tests for categorical variables and one-way ANOVA for continuous variables. Normalized gene expression counts of 16 was set as the lower limit of detection and signal lower than the LLD was set at 16 prior to data analysis. Distribution of the gene expression of the genes was studied with scatterplots. The gene expression of TLR1, TLR6, TLR5, MAPK14, CCR1 and ICAM1 were divided into quartiles and the highest quartile was divided into two levels: 75th to 95th percentile and>95th to 100th percentile (the top twentieth). We evaluated the association between lung function at CARDIA exam years 30, 10-year decline in lung function (year 20-year 30) and year 25 gene expression levels of TLR5, MAPK14 and CCR1 using linear regression models after adjustment for center, smoking status (never, former, current smokers at year 25) and body mass index (BMI) (year 25). We defined percent predicted lung function as the ratio of observed lung function over predicted lung function, where predicted lung function was calculated using the Hankinson equation30. We created an inflammation score by adding the quartile levels of TLR5, MAPK14 and CCR1 for each participant so that the maximum inflammation score would be 15 when all markers were in the highest level (top twentieth) while the minimum inflammation would be 3 when all markers were in the first quartile. We also created an inflammation score using quartile levels of the six genes such that the maximum inflammation score is 30 when the six genes are in the highest level and the minimum inflammation score is 6 when the six genes are in the first quartile. Multivariable linear regression models were used for evaluating the association between lung function at CARDIA exam year 30, 10-year decline in lung function and inflammation score as a continuous predictor variable. Sensitivity analysis was performed by excluding asthma and COPD patients at CARDIA exam years 20 and 30, and evaluation of the association between lung function at CARDIA exam year 30, 10- year decline in lung function and year 25 gene expression levels of TLR1, TLR5, TLR6, MAPK14,CCR1, ICAM1 in the subgroup of participants without COPD/asthma. All the p-values ≤ 0.05 were considered statistically significant. Statistical analyses were carried out using SAS software version 9.4 (SAS Institute, Cary, NC).
Results
Characteristics at year 25 examination
The participants in highest level (top twentieth) of TLR5 at year 25 were more likely to be black (57.52% vs 44.64%; p-value = 0.0006), current smokers (23.01% vs. 8.80%; p < 0.0001), have higher BMI (34.34 kg/m2 vs. 28.66 kg/m2; p < 0.0001) and have higher C-reactive protein (CRP) (7.79 µG/mL vs. 1.69 µG/mL; p < 0.0001) as compared to those in the first quartile of TLR5 (Table 1).There were no consistent differences in age and alcohol consumption among the five levels of TLR5. In general, participants with the highest levels of other genes such as CCR1, TLR1, TLR6, and ICAM1 showed similar distribution as TLR5 with participants being more likely to be black, women, have higher BMI and higher CRP as compared to the respective genes’ first quartiles, except for MAPK14, where the participants in the highest level of MAPK14 were more likely to be white (63.72% vs. 39.08%; p-value <0.001). (Supplementary Tables 1a-1e). The scatterplots of the gene expressions showed a wide range of the distribution of the gene expression levels for all six genes, with and highest correlation seen between TLR1 and TLR6 (r = 0.73; p- <0.01). In general, there was a higher correlation between the TLR genes (TLR1, TLR5 and TLR6) (r = 0.49–0.73; p < 0.01) while there were only modest correlations seen between TLR genes and genes in downstream pathways such as CCR1 and ICAM1 (r = 0.23–0.33; p < 0.01). However, the associations between TLR genes and MAPK14 were stronger (r = 0.54–0.64; p < 0.01) (Supplementary Fig. 1).
Association between year 30 lung function and year 25 gene expression profiles
Year 30 predicted FEV1 and FVC was lower in the highest level (top twentieth) of TLR5 as compared to the lowest quartile of TLR5, with a difference of 4.00% (95% CI: 0.95–7.05; p for trend: 0.04) and 3.90% (95% CI: 1.14–6.65; p for trend: 0.05), respectively. (Table 2). None of the other genes were associated with year 30 FEV1 or FVC (Table 2, Supplementary Table 2). Inflammation score was associated with year 30 predicted FEV1 and FVC. The difference in FEV1 between the highest inflammation score (for TLR5, MAPK14 and CCR1) and lowest inflammation score was 6.21 (95% CI: −2.25, 14.68; p = 0.02) while difference in FVC was 5.34 (95% CI: −2.34, 13.02; p = 0.03). Year 30 predicted FEV1/FVC and the inflammation score were not associated with FEV1/FVC (Table 2, Supplementary Table 2). The inflammation score using six genes was not associated with year 30 FEV1, FVC and FEV1/FVC ratio. The difference between year 30 predicted FEV1 and FVC at maximum inflammation score compared to minimum inflammation score was 8.68% (95% CI: −13.81, 31.18) and 9.87 (95% CI: −10.46, 30.19), respectively.
Association between 10-year decline in lung function and year 25 gene expression profiles
Decline in FEV1, FVC and FEV1/FVC from year 20 to year 30 was not significantly associated with year 25 TLR5 expression levels (Table 3). Contrary to our hypothesis, decline in FEV1 and FVC was lower in the highest level (top twentieth) of MAPK14 as compared to the lowest quartile (1.60% vs 3.19%; p for trend =0.03 and 1.86% vs 3.73%; p for trend=0.008). Decline in FEV1/FVC was not significantly associated with MAPK14 expression levels. Consistent with our hypothesis, decline in FEV1 was significantly higher in the highest level of CCR1 as compared to the lowest quartile (3.43% vs. 1.73%; p for trend=0.01) (Table 3). Decline in FVC and FEV1/FVC was not significantly associated with CCR1. Expression levels of other genes were not associated with 10-year decline in FVC, FEV1 and FEV1/FVC (Supplementary Table 3). The inflammation score of TLR5, CCR1 and MAPK14 was not associated with the 10-year decline in FEV1, FVC and the FEV1/FVC ratio (difference in 10 year decline in FEV1 between highest and lowest inflammation scores = 1.11% (95% CI: −3.95,6.16; p = 0.83), difference in 10 year decline in FVC between highest and lowest inflammation scores = −0.85 (95% CI: −5.86, 4.16; p = 0.52), difference in 10 year decline in FEV1/ FVC between highest and lowest inflammation scores = 2.03 (95% CI: −0.88, 4.94; p = 0.39). The inflammation score created using six genes was not associated with 10-year decline of FEV1, FVC and FEV1/FVC ratio. The difference between 10-year decline of predicted FEV1 and FVC at maximum inflammation score compared to minimum inflammation score was −5.53 (95% CI: −24.11, 13.05) and −9.33 (95% CI: −27.74, 9.07), respectively.
Sensitivity analysis after exclusion of asthma and COPD patients
The distribution of asthma and COPD participants was similar across all quartiles of MAP14 (24.29% vs. 20.37%; p = 0.32 and 3.06% vs. 2.43%; p = 0.82), CCR1 (24.66% vs. 19.10%; p = 0.07 for asthma and 3.29% vs 2.71%; p = 0.49 for COPD). The distribution across TLR5 quartiles was different (26.30% vs. 20.03%; p = 0.02 for asthma and 4.33% vs. 1.52% for COPD; p = 0.01). After removal of the asthma and COPD patients, we found results similar to those seen in the entire dataset. Briefly, year 30 FEV1 and FVC was lower in the highest level (top twentieth) of TLR5 compared to the lowest level, with a difference of 4.84 (95% CI: 1.68–8.01; p for trend: 0.05) and 4.83 (95%CI: 1.87–7.78; p for trend: 0.03), respectively. The 10-year decline of FEV1 was higher in the highest level of CCR1 as compared to the lowest level with as difference of 2.76 (95% CI: 0.68–4.83; p for trend: 0.02). The 10-year decline of FEV1 and FVC was lower in the highest level of MAPK14 as compared to lowest level with a difference of 1.63% (95% CI: −0.41–3.69; p for trend: 0.01) and 2.01% (95% CI: −0.06–4.08; p for trend: 0.006).
Discussion
This study showed that higher expression of TLR5 was associated with lower year 30 predicted FEV1 and FVC. In addition, faster 10-year decline of FEV1 from year 20 to year 30 was associated with higher gene expression levels of CCR1. However, the faster 10-year decline in predicted FEV1 and FVC was associated with lower year 25 gene expression levels of MAPK14. These results are consistent with the hypothesis that higher gene expression levels of genes in the pathogen recognition pathways were associated with lower lung function for the most part.
Previous studies on inflammation and lung function have utilized non-specific markers of inflammation2,4,31,32 and, to our knowledge, this is the first study to evaluate and demonstrate an inverse association between gene expression levels in the pathogen recognition pathway and lung function. A study done with 531 participants from the European Community Respiratory Health Survey found a cross sectional negative relationship between percent predicted FEV1 and serum CRP concentration after dividing them into tertiles (p-value = 0.002). Other studies which used markers of systemic inflammation such as fibrinogen also reported a faster lung function decline was associated with higher fibrinogen levels31,33. With respect to longitudinal relationship, there have been mixed findings with CRP. A previous study in CARDIA reported a faster decline in FEV1 and FVC from year 5 to year 20 among participants in the highest quartile of year 7 CRP compared to the lowest quartile4. The study conducted with participants from the European Community Respiratory Health Survey also found that increase in CRP levels was associated with greater mean annual FEV1 decline after adjustment for potential confounders (p-value = 0.002)34. However, another community based cohort with 2442 individuals found that the baseline CRP value was not associated with FEV1 and FVC decline over 9 years32. These studies suggest that an overall increase in inflammation is associated with reduced lung function but do not indicate a specific pathway leading to this increase.
Results observed in our study suggest that higher levels of gene expression of a specific inflammatory pathway, the pathogen recognition pathway, is associated with lower lung function. TLR5 is a cell surface marker which recognizes flagellin and activates proinflammatory markers. TLR5 mediated signaling in the airway structural cells, following exposure to flagellin, triggers compartmentalized mucosal innate immunity and results in improved T-cell-mediated immunity and antibody responses35,36. A previous study identified flagellin as a microbial product that stimulated strong allergic responses and promoted allergic sensitization to indoor allergens, by stimulating secretion of pro-inflammatory cytokines by lung epithelial cells23. Another study has shown that TLR5 signaling in the airway epithelium is important for induction of proinflammatory responses such as chemokine production in neutrophils and macrophages37. Consistent with these studies, the present study showed that TLR5 gene expression levels were associated with year 30 predicted FEV1 and FVC values suggesting that pathogen recognition maybe an important pathway associated with lung function. We hypothesize that TLR5 overexpression may lead to excessive proinflammatory responses in the lung that ultimately reduce lung function. Since TLR5 was associated with lower lung function but not lung function decline over 10 years, our results suggest that TLR5 may be important in determining early life lung function and influencing the peak lung function attained rather than impact decline in lung function among adults. This study also showed that higher levels of CCR1, a downstream marker in the TLR2/TLR1 pathogen recognition pathway, was associated with faster decline of FEV1. Asthmatics have higher CCR1 expression in airway smooth muscle cells as compared to healthy controls38 and another study found increased expression of CCR1 positive mast cells in bronchial biopsies in asthmatic patients compared to healthy controls39. These studies indicate that overexpression of CCR1 may play an important role in asthma and reduced lung function. Since CCR1 was associated with lung function decline in our study, we hypothesize that CCR1 may have a stronger influence on decline of lung function in adults. Thus, TLR5 and CCR1 may influence lung function during different timepoints and overexpression of both genes may have a synergistic effect on reducing lung function. Contrary to our hypothesis, we found that 10-year decline of FEV1 and FVC was slower in higher levels of MAPK14. This needs to be investigated further as the reasons for this finding remains unclear.
The results observed in our study suggest that only extremely high levels of the markers in the TLR pathway reduce lung function suggesting biological resilience to small changes of gene expression levels in individual biological pathways. For example, the difference in lung function between the fourth and the fifth levels of FEV1 and FVC at year 30 for TLR5 was higher than the difference between the other levels. The lower values of FEV1 and FVC at the highest 5% levels of the markers suggests that there may be a threshold value of gene expression levels of these markers influencing the decrease in lung function. However, the higher difference between fourth and fifth levels is not observed in the analysis between decline in lung function and levels of inflammatory markers. Lower FEV1 and FVC, within the normal range, have been shown to be strong predictors of overall mortality and other health outcomes such as cardiovascular diseases40,41,42,43. Previous studies44,45, have also shown that decline in FEV1 or FVC with preserved ratio is predictive of cardiovascular disease, and that this “restrictive” condition is associated with a “Hypertrophic, high output cardiac phenotype”, while obstructive disease with declining FEV1/FVC ratio is associated with a “Small heart, low output phenotype”40. While the association between decline in FEV1 or FVC and chronic respiratory disease has not been evaluated, we hypothesize that lower FEV1 and FVC and/or faster declines in FEV1 or FVC, within the normal range, may represent an intermediate phase between ideal lung health and chronic respiratory disease46. This and other studies conducted in the CARDIA cohort, provide a foundation to develop a model that informs us of transition from normal lung health to chronic respiratory disease, and study early changes in lung function as a risk factor for pulmonary and cardiovascular health outcomes.
The strengths of the study include the long-term follow-up of the participants and the representative sample with inclusion of blacks and whites, and men and women. Assessing the markers using gene expression analysis is advantageous in cases where measurement of protein level is not possible. The limitations of the study include the timing of the measurements of lung function and the gene expression markers at different years thereby limiting our understanding of the temporal relationship between gene expression levels and lung function. Measurement of gene expression levels at more than one interval will help understanding the longitudinal relationship further. However, gene expression levels measured at year 25 were analyzed with FEV1 and FVC measured at year 30, aligning the temporality with our hypothesis about the effect of gene expression on lung function measures. Measurement of FEV1 and FVC using different methods at year 20 and year 30 (a dry rolling-seal OMI spirometer at year 20 and a portable spirometer at year 30) could have impacted the measurements. However, we have followed ATS guidelines for measurement of lung function at both times thereby minimizing the variation in lung function measurements across both visits. Gene expression levels of these inflammatory markers could be correlated with differences in cell composition such as the proportion of monocytes, T-lymphocytes etc. Since complete blood counts are not available in CARDIA at year 25, differences in cell composition may be a potential confounder in the observed association. A study evaluating the expression of Toll like receptors on peripheral blood mononuclear cells (PBMCs) showed that TLR5 was expressed in T cells, NK cells and monocytes but was not expressed in B cells or plasmacytoid dendritic cells.47. In addition, TLR5 is also expressed in airway neutrophils48. Thus, TLR5 appears to be expressed in all major cell subsets in peripheral blood apart from the B cells. Thus, changes in B cell distribution may impact TLR5 expression in peripheral blood. Lastly, we did not adjust for multiple comparisons in these analyses as these genes were selected based on a a priori hypothesis regarding the role of inflammatory markers in lung function. Using Bonferroni correction and a p-value of 0.003 (6 biomarkers with three outcomes = 0.05/18 = 0.0003) to determine statistical significance would result in none of the observed associations being statistically significant. Hence these results, though suggestive of an association between gene expression levels in TLR5, MAPK14 and CCR1 and lung function, need to be confirmed in independent studies.
In conclusion, the results suggest that high levels of gene expression of TLR5 and CCR1 are associated with lower lung function and these results are independent of smoking and BMI. These results suggest that pathogen recognition pathway may be important in influencing lung health. Future studies that include measurement of gene expression levels at multiple timepoints in independent datasets need to be conducted to determine the specific genes that may be important in the longitudinal decline in lung function in healthy adults and whether modulation of the pathogen recognition pathways may be helpful in improving lung health among young adults.
References
Cho, Y. S. & Moon, H.-B. The Role of Oxidative Stress in the Pathogenesis of Asthma. Allergy. Asthma Immunol. Res. 2, 183–187 (2010).
Hancox, R. J. et al. Systemic inflammation and lung function in young adults. Thorax 62, 1064–1068 (2007).
Dahl, M., Tybjaerg-Hansen, A., Vestbo, J., Lange, P. & Nordestgaard, B. G. Elevated plasma fibrinogen associated with reduced pulmonary function and increased risk of chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 164, 1008–1011 (2001).
Kalhan, R. et al. Systemic Inflammation in Young Adults Is Associated with Abnormal Lung Function in Middle Age. PLoS One 5(7), e114 (2010).
Rahman, I. The role of oxidative stress in the pathogenesis of COPD: implications for therapy. Treat. Respir. Med. 4, 175–200 (2005).
Schunemann, H. J. et al. Oxidative stress and lung function. Am. J. Epidemiol. 146, 939–948 (1997).
Black, S., Kushner, I. & Samols, D. C-reactive Protein. J. Biol. Chem. 279, 48487–48490 (2004).
Shrivastava, A. K., Singh, H. V., Raizada, A. & Singh, S. K. C-reactive protein, inflammation and coronary heart disease. Egypt. Hear. J. 67, 89–97 (2015).
Tang, W. et al. Genetic Associations for Activated Partial Thromboplastin Time and Prothrombin Time, their Gene Expression Profiles, and Risk of Coronary Artery Disease. Am. J. Hum. Genet. 91, 152–162 (2012).
Mendelson, M. M. et al. Association of Body Mass Index with DNA Methylation and Gene Expression in Blood Cells and Relations to Cardiometabolic Disease: A Mendelian Randomization Approach. PLoS Med 14 (2017).
Janssens, S. & Beyaert, R. Role of Toll-Like Receptors in Pathogen Recognition. Clin. Microbiol. Rev. 16, 637–646 (2003).
Iwasaki, A. & Medzhitov, R. Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 5, 987 (2004).
Bezemer, G. F. G. et al. Dual Role of Toll-Like Receptors in Asthma and Chronic Obstructive Pulmonary Disease. Pharmacol. Rev. 64, 337–358 (2012).
Phipps, S., Lam, C. E., Foster, P. S. & Matthaei, K. I. The contribution of toll-like receptors to the pathogenesis of asthma. Immunol. Cell Biol. 85, 463 (2007).
Jin, M. S. et al. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell 130, 1071–1082 (2007).
Oliveira-Nascimento, L., Massari, P. & Wetzler, L. M. The Role of TLR2 in Infection and Immunity. Front. Immunol. 3, 79 (2012).
Neote, K., DiGregorio, D., Mak, J. Y., Horuk, R. & Schall, T. J. Molecular cloning, functional expression, and signaling characteristics of a C-C chemokine receptor. Cell 72, 415–425 (1993).
Parker, L. C., Whyte, M. K. B., Vogel, S. N., Dower, S. K. & Sabroe, I. Toll-Like Receptor (TLR)2 and TLR4 Agonists Regulate CCR Expression in Human Monocytic Cells. J. Immunol. 172, 4977–4986 (2004).
Thyagarajan, B. et al. Association of Circulating Adhesion Molecules With Lung Function: The CARDIA Study. Chest 135, 1481–1487 (2009).
Mudaliar, H. et al. The Role of TLR2 and 4-Mediated Inflammatory Pathways in Endothelial Cells Exposed to High Glucose. PLoS One 9, e108844 (2014).
Yoon, S. et al. Structural basis of TLR5-flagellin recognition and signaling. Science 335, 859–864 (2012).
Smith, K. D. et al. Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat. Immunol. 4, 1247 (2003).
Wilson, R. H. et al. The TLR5 ligand flagellin promotes asthma by priming allergic responses to indoor allergens. Nat. Med. 18, 1705–1710 (2012).
Yu, Y. et al. TLR5-mediated activation of p38 MAPK regulates epithelial IL-8 expression via posttranscriptional mechanism. Am. J. Physiol. Liver Physiol. 285, G282–G290 (2003).
Farhat, K. et al. Heterodimerization of TLR2 with TLR1 or TLR6 expands the ligand spectrum but does not lead to differential signaling. J. Leukoc. Biol. 83, 692–701 (2008).
Ono, K. & Han, J. The p38 signal transduction pathway Activation and function. Cell. Signal. 12, 1–13 (2000).
Hughes, G. H. et al. Recruitment in the Coronary Artery Disease Risk Development in Young Adults (Cardia) Study. Control. Clin. Trials 8, 68S–73S (1987).
Friedman, G. D. et al. CARDIA: study design, recruitment, and some characteristics of the examined subjects. J. Clin. Epidemiol. 41, 1105–1116 (1988).
Miller, M. R. et al. Standardisation of spirometry. Eur. Respir. J. 26, 319 LP-338 (2005).
Hankinson, J. L., Odencrantz, J. R. & Fedan, K. B. Spirometric reference values from a sample of the general U.S. population. Am. J. Respir. Crit. Care Med. 159, 179–187 (1999).
Thyagarajan, B. et al. Plasma fibrinogen and lung function: the CARDIA Study. Int. J. Epidemiol. 35, 1001–1008 (2006).
Fogarty, A. W., Jones, S., Britton, J. R., Lewis, S. A. & McKeever, T. M. Systemic inflammation and decline in lung function in a general population: a prospective study. Thorax 62, 515–520 (2007).
Donaldson, G. C. et al. Airway and Systemic Inflammation and Decline in Lung Function in Patients With COPD. Chest 128, 1995–2004 (2005).
Shaaban, R. et al. Change in C-reactive protein levels and FEV1 decline: a longitudinal population-based study. Respir. Med. 100, 2112–2120 (2006).
Van Maele, L. et al. Airway structural cells regulate TLR5-mediated mucosal adjuvant activity. Mucosal Immunol. 7, 489 (2013).
Malash, A. H., Ali, A. A., Samy, R. M. & Shamma, R. A. Association of TLR polymorphisms with bronchopulmonary dysplasia. Gene 592, 23–28 (2016).
Parker, D. P. A. Epithelial Uptake of Flagella Initiates Proinflammatory Signaling. PLoS One 8(3), e599 (2013).
Joubert, P. et al. Expression and Regulation of CCR1 by Airway Smooth Muscle Cells in Asthma. J. Immunol. 180, 1268 LP–1275 (2008).
Amin, K., Janson, C., Harvima, I., Venge, P. & Nilsson, G. CC chemokine receptors CCR1 and CCR4 are expressed on airway mast cells in allergic asthma. J. Allergy Clin. Immunol. 116, 1383–1386 (2005).
Cuttica, M. J. et al. Loss of Lung Health from Young Adulthood and Cardiac Phenotypes in Middle Age. Am. J. Respir. Crit. Care Med. 192, 76–85 (2015).
Jacobs, D. R. Jr et al. Rate of decline of forced vital capacity predicts future arterial hypertension: the Coronary Artery Risk Development in Young Adults Study. Hypertens. (Dallas, Tex. 1979) 59, 219–225 (2012).
Cuttica, M. J. et al. Lung Function in Young Adults and Risk of Cardiovascular Events Over 29 Years: The CARDIA Study. J. Am. Heart Assoc. 7, e010672–e010672 (2018).
Jacobs, D. R. Jr & Kalhan, R. Healthy Diets and Lung Health. Connecting the Dots. Ann. Am. Thorac. Soc. 13, 588–590 (2016).
Kannel, W. B., Hubert, H. & Lew, E. A. Vital capacity as a predictor of cardiovascular disease: the Framingham study. Am. Heart J. 105, 311–315 (1983).
Engström, G., Melander, O. & Hedblad, B. Population-based study of lung function and incidence of heart failure hospitalisations. Thorax 65, 633–638 (2010).
Reyfman, P. A. et al. Defining Impaired Respiratory Health. A Paradigm Shift for Pulmonary Medicine. Am. J. Respir. Crit. Care Med. 198, 440–446 (2018).
Hornung, V. et al. Quantitative Expression of Toll-Like Receptor 1–10 mRNA in Cellular Subsets of Human Peripheral Blood Mononuclear Cells and Sensitivity to CpG Oligodeoxynucleotides. J. Immunol. 168, 4531 LP–4537 (2002).
Koller, B. et al. TLR expression on neutrophils at the pulmonary site of infection: TLR1/TLR2-mediated up-regulation of TLR5 expression in cystic fibrosis lung disease. J. Immunol. 181, 2753–2763 (2008).
Acknowledgements
The CARDIA study is supported by contracts HHSN268201300025C, HHSN268201300026C, HHSN268201300028C, HHSN268201300029C, and HHSN268200900041C from the National Heart, Lung, and Blood Institute (NHLBI); the Intramural Research Program of the National Institute on Aging (NIA); and an intra‐agency agreement between the NIA and NHLBI (AG0005) and grant R01 HL122477 (to Kalhan). This manuscript has been reviewed by CARDIA for scientific content.
Author information
Authors and Affiliations
Contributions
R.R. worked on the data analysis and drafted the manuscript; R.K., D.J., L.H., G.W. and B.T. helped with the critical review of the analysis and manuscript; M.G. conducted the gene expression measurement experiments; W.G. conducted the data analysis for the gene expression measurements.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ramasubramanian, R., Kalhan, R., Jacobs, D.R. et al. Lung Function and Gene Expression of Pathogen Recognition Pathway Receptors: the Cardia Lung Study. Sci Rep 10, 9360 (2020). https://doi.org/10.1038/s41598-020-65923-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-65923-z
- Springer Nature Limited