Abstract
This article describes primary data and resources available from the Boston Adolescent Neuroimaging of Depression and Anxiety (BANDA) study, a novel arm of the Human Connectome Project (HCP). Data were collected from 215 adolescents (14–17 years old), 152 of whom had current diagnoses of anxiety and/or depressive disorders at study intake. Data include cross-sectional structural (T1- and T2-weighted), functional (resting state and three tasks), and diffusion-weighted magnetic resonance images. Both unprocessed and HCP minimally-preprocessed imaging data are available within the data release packages. Adolescent and parent clinical interview data, as well as cognitive and neuropsychological data are also included within these packages. Release packages additionally provide data collected from self-report measures assessing key features of adolescent psychopathology, including: anxious and depressive symptom dimensions, behavioral inhibition/activation, exposure to stressful life events, and risk behaviors. Finally, the release packages include 6- and 12-month longitudinal data acquired from clinical measures. Data are publicly accessible through the National Institute of Mental Health Data Archive (ID: #2505).
Similar content being viewed by others
Background & Summary
Adolescence is a critical time for understanding brain changes associated with psychiatric disorders. The brain’s emotion-, executive-, and reward-related circuitries undergo significant development during adolescence1,2. These circuits are also implicated in the etiology and maintenance of the most common forms of psychopathology, anxiety and depression3,4,5. Here, we describe data and resources available from the Boston Adolescent Neuroimaging of Depression and Anxiety (BANDA) study, which provides the first ever large, public-access neuroimaging dataset specifically targeting adolescent anxiety and depression.
Other large neuroimaging initiatives have targeted brain development, such as the Adolescent Brain and Cognitive Development study6, IMAGEN7, Philadelphia Neurodevelopmental Cohort8, and the Human Connectome Project lifespan development study (HCP-D)9,10. The BANDA study, however, was innovative in its focus on adolescent anxiety and depression. Specifically, BANDA oversampled adolescents with at least one of these disorders (71% of participants) and acquired both neuroimaging (i.e., task-based fMRI) and clinical measures relevant to anxiety and depression. As part of the broader Connectomes Related to Human Diseases initiative, BANDA MRI data were acquired via sequences harmonized with the HCP and imaging data released underwent HCP quality control and minimal preprocessing by the Connectome Coordination Facility11,12. These procedures were intended to enhance the reproducibility of findings from BANDA data, lessen the computational requirements for end users, as well as provide the means for standardized comparisons with other HCP studies. Further, to facilitate cross-study comparisons, many BANDA measures were specifically harmonized with the HCP-D and other related Connectomes Related to Human Diseases studies13.
Here, we describe the primary MRI, clinical, and neurocognitive data from the BANDA study. In total, data from 215 adolescents (imaging acquired at ages 14–17, Mean = 15.4 years) were collected; 152 of whom had a current diagnosis of an anxious and/or depressive disorder. The current data release packages can be accessed through the National Institute of Mental Health Data Archive (NDA; ID #2025).
Methods
Primary data collection began October 2016 and lasted until November 2021. This acquisition protocol entailed four sessions. Participating parents provided informed consent and adolescents assented to study procedures and data sharing. Study procedures and data sharing were approved by the Massachusetts General Brigham Institutional Review Board (Protocol #P002534). Parents and adolescents were compensated for their time.
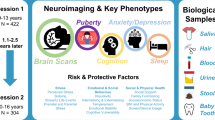
Two broad categories of data were collected, non-MRI and MRI data. Non-MRI data were acquired during Sessions 1–4, many of which were from repeated measures to permit longitudinal analyses. MRI data were cross-sectional and solely acquired during Session 2.
Schedule and criteria
Participant recruitment and screening
Participants were recruited from clinics, mass transit advertisements, newsletters to special interest groups, and social media. Interested parents and adolescents were screened via phone for inclusion/exclusion criteria.
Clinical and neurocognitive characterization (Session 1)
Participants meeting initial inclusion criteria were enrolled in the study and underwent clinical/neuropsychological testing at one of three affiliated clinical sites: the Center for Anxiety and Related Disorders at Boston University; the Center for Depression, Anxiety, and Stress Research at McLean Hospital/Harvard Medical School; or the Child Cognitive Behavioral Therapy Program at Massachusetts General Hospital/Harvard Medical School. Clinical diagnoses and neuropsychological metrics obtained during Session 1 were used to confirm inclusion/exclusion criteria. Briefly, adolescents included were ages 14–17 at the time of imaging; fluent in English; able and willing to undergo MRI; and had an intelligence quotient (IQ) score of \(\ge \)85. This latter inclusion criterion was relaxed during the study and adolescents were allowed to matriculate based upon the experimenter’s clinical judgement. Parents were also fluent in English and needed to have an IQ of \(\ge \)85. Exclusion criteria were: premature birth (<37, or <34 weeks for twins) or less than 5 lbs at birth; serious medical condition; history of serious head injury; hospitalization for >2 days for neurovascular or cardiovascular disease; diagnosis of autism spectrum disorder; daily use of migraine medication or migraine within 72 hours of scanning.
Brain imaging (Session 2)
Participants meeting inclusion criteria underwent brain imaging at the Massachusetts General Hospital Athinoula A. Martino’s Center for Biomedical Imaging at Harvard Medical School. Efforts were made to minimize the interval between imaging and Session 1, with 65.7% of participants scanned within 1–2 weeks of Session 1, 94.6% within 1–4 weeks, and 97.0% within 1–5 weeks.
Longitudinal clinical data collection (Sessions 3, 4)
Longitudinal data on clinical measures were also acquired at two, 6-month intervals. Session 3 occurred online, approximately 6 months after brain imaging and included a second acquisition of adolescent self-report measures obtained from Session 1. Session 4 occurred approximately 12 months after brain imaging. This session was designed to be completed in person. However, due to local shelter-in-place orders and moratoriums on in-person data collection during the COVID-19 pandemic, some participants completed Session 4 remotely (e.g., via videoconference). Session 4 consisted of (1) a third acquisition of adolescent self-report measures; (2) a second acquisition of adolescent and parent structured clinical interviews; and (3) a second acquisition of parent-report clinical measures (report on self and on adolescent).
Non-MRI characterization
For brevity, most of the names of non-MRI measures are listed in Fig. 3c.
Structured diagnostic interviews and neuropsychological tests
Each adolescent and their participating parent were interviewed by trained staff who were, or were under the supervision of, a licensed clinical psychologist. Adolescents and parents were separately administered a structured clinical interview to assess present and lifetime psychiatric disorders for the adolescents (Kiddie-SADS)14. This interview was amended by study psychologists to assess DSM-5 criteria15. Family history of psychiatric disorders was assessed via a structured interview with the participating parent16. Suicide risk of adolescents was assessed via the Columbia Suicide Severity Rating Scale17. Each adolescent and their participating parent were administered an abridged standardized intelligence test18.
Data were recorded on standardized interviewer forms14,16,17,18 and then manually entered into a secure web-based application (i.e., REDCap)19. To ensure manually entered data faithfully reflected participant responses and the interviewers’ clinical impressions, most of these data underwent a three-tier accuracy assurance protocol. In the first tier, data were manually entered into REDCap from interviewer notes by a staff member at an affiliated clinical site and by a separate staff member at a different site. In the second tier, reliability of these entries was assessed algorithmically. Any discrepancies were redressed in the third tier by re-examining the interviewer notes, obtaining verbal confirmation regarding the intention of the clinical interviewer, and/or listening to an audio file of the clinical interview. Accuracy assured, item-level, raw interview data were then transformed into relevant NDA elements.
Group/Phenotype labels
One of three group (i.e., phenotype) labels were ascribed to each participant based upon interviews and clinical impressions obtained during Session 1. Thus, these labels only pertained to study baseline/intake. These labels included: Anxiety, Depression, or Control. The Anxiety label was assigned to 85 adolescents (39.5%) who did not meet criteria for any current depressive disorder, but who met DSM-5 criteria for at least one anxious disorder that was either in partial remission or clinically definite, including: agoraphobia, generalized anxiety, panic disorder, separation anxiety disorder, specific phobia, or other unspecified anxiety disorder. The Depression label was assigned to 67 adolescents (31.2%) who met DSM-5 criteria for at least one depressive disorder that was either in partial remission or clinically definite, including: adjustment disorder with depressed mood, depressive disorder unspecified (NOS), dysthymia, or major depressive disorder. Note that a current anxious disorder was not exclusionary for the Depression label and 56 adolescents (83.6%) in this phenotype group met criteria for at least one comorbid anxiety disorder (i.e., agoraphobia, generalized anxiety disorder, panic disorder, separation anxiety disorder, social anxiety disorder, specific phobia) at Session 1. The Control label was assigned to 63 (29.3%) adolescents who did not meet current criteria for a DSM-5 disorder nor did they meet lifetime criteria for an anxious or depressive disorder. The DSM-5 diagnoses that were used to assign the Anxiety, Depression, and Control labels were evaluated for inter-rater agreement using an initial, stratified random sample of BANDA adolescents and a blinded, licensed clinical psychologist. Inter-rater agreement ranged from moderate (anxiety diagnoses, \(\kappa \) = 0.55) to substantial (depression diagnoses, \(\kappa \) = 0.66) and exceeded levels of agreement attained by the initial field trials of the DSM-5 itself20, as detailed in our prior report21.
Adolescent and parent report
Adolescents were administered 8 self-report measures to assess relevant clinical symptoms and behavioral characteristics. Adolescents were also administered a questionnaire on their physical development22 and hand dominance23. Parents were administered three self-report measures to characterize dimensions of the adolescents’ moods, personality traits, and behaviors. Parents were also administered two self-report measures describing their own moods, personality traits, thoughts, and behaviors. Demographic data were acquired via parent report, along with data on adolescents’ current psychiatric medications.
Data for each self-report measure were collected via computer and sent to a secure, REDCap server. All data were sent directly to a study REDCap server, except those from the adolescent Stress and Adversity Inventory (STRAIN)24. STRAIN data were initially transmitted to a secure server at the University of California, Los Angeles (UCLA). These item-level, raw self-report data were transformed to composite-level data by the STRAIN-UCLA team and then transferred to the study REDCap server. All available item-level, raw self-report data were transformed into relevant structures and exported to the NDA. Select summary/composite scores were also created consistent with extant element structures and transferred to the NDA.
Computerized cognitive testing
Adolescents received 9 computerized cognitive tests from two standardized batteries; the NIH Toolbox (version 1.11)25,26 and the University of Pennsylvania Computerized Neuropsychological Test Battery (Penn Test Battery27). These measures assess various domains including: cognitive flexibility/attention (NIH Toolbox Dimensional Change Card Sort Test); emotion recognition (Penn Emotion Recognition Task); inhibition/attention (NIH Toolbox Flanker Task); impulsivity/self-regulation (Penn Delay Discounting Task); nonverbal reasoning (Penn Matrix Reasoning Test); processing speed (NIH Toolbox Pattern Comparison Processing Speed Test); working memory (NIH Toolbox List Sorting Working Memory Test); reading decoding (NIH Toolbox Oral Reading Recognition Test); verbal episodic memory (Penn Word Memory Test).
NIH Toolbox tests were administered on iPads (iOS versions 9.3.2 and 11.2.6). Transformation of NIH Toolbox raw data to composite-level data occurred automatically within the iPad application. Details of these transformations are found elsewhere28. Both composite-level and raw data from NIH Toolbox measures were sent to a secure study server. Composite-level data were then exported to the NDA. Penn Task Battery raw data were transmitted to a secure University of Pennsylvania server, composite-level data were then transferred back to the study’s secure server. Where necessary, data were transformed into relevant structures before exporting to the NDA.
MRI characterization
Images
Descriptions and findings pertaining to the harmonization of imaging sequences with the HCP are provided elsewhere13. Sequences were based upon the HCP Lifespan protocols. As part of licensing agreements, sequences were updated during the course of this study.
MRI data were collected via a 3 Tesla Siemens Prisma and a 64-channel head coil. T1- and T2-weighted images were acquired with a 0.8 mm isotropic voxel resolution (collection aliases: T1w_MPR_vNav and T2w_SPC_vNav). T1- and T2-weighted acquisitions leveraged volumetric navigators (vNavs) for prospective motion correction29. Four runs of diffusion-weighted images were acquired in an equal number of alternating phase-encoding directions (collection alias: dMRI). These featured a 1.5 mm isotropic voxel resolution, b-values of 1500 and 3000 s/mm2, and 183 diffusion directions. Blood-oxygen-level dependent (BOLD) images were acquired with an 800 ms TR and 2 mm isotropic spatial resolution. BOLD images were also acquired in alternating phase-encoding directions (Anterior Posterior – AP and PA). To facilitate post-acquisition corrections of susceptibility artifacts, spin-echo sequences were acquired proximal to the beginning of each imaging run. Four runs (420 frames = 5 min 46 s per run) of BOLD images were acquired while participants were asked to rest quietly with their eyes open while a fixation cross was placed on the screen (i.e., resting state; collection alias: rfMRI_REST). BOLD images were also acquired while participants completed three tasks, described here briefly and elsewhere in greater detail21.
Incentive processing task (IPT, 2 runs, 215 frames per run)
Collection alias: tfMRI_GAMBLING. The IPT is a block-design task that presented adolescents with the prospect of monetary gains or losses30,31. Participants were instructed to guess whether a forthcoming number was greater or less than 5 on each trial. A block lasted 28 s and involved 8 trials wherein participants were given mostly reward feedback or mostly loss feedback on their guess (Fig. 1a). Four reward and four loss blocks were presented per each of the two runs.
Emotion processing task (EPT, 2 runs, 338–405 frames per run)
Collection alias: tfMRI_FACEMATCHING. The EPT is a block-design task that presented adolescents with images of faces or objects32,33. A trial consisted of three images from one of five conditions, including faces with actor’s portraying either fearful, happy, sad, or neutral expressions or object images. Participants were instructed to determine which of two images presented at the bottom of the frame matched a single image presented at the top (Fig. 1b). Blocks lasted 18 s and involved 6 trials from the same condition. Three blocks for each condition were presented per each of the two runs.
Emotion interference task (EIT, 4 runs, 280 frames per run)
Collection alias: tfMRI_CONFLICT. The EIT is an event-related task that presented adolescents with pictures of faces and houses in orthogonal visuospatial axes. Participants were cued to attend to a given visuospatial axis which featured images of either faces or houses, and ignore images presented on the orthogonal axis34,35,36. For each trial, participants were instructed to indicate whether two images presented on the cued axis were identical or different. Face images consisted of actors portraying either fearful or neutral expressions. Trials began with a fixation cross (1 s) which was followed by the presentation of the images (0.25 s). Then, participants were allotted 2.2 s for their response. Event conditions can be constructed in several ways, including a four-condition model: attend fearful faces (ignore houses), attend neutral faces (ignore houses), ignore fearful faces (attend houses), and ignore neutral faces (attend houses). Within this construction, each of the four conditions were presented 24 times across the four task runs. A simpler, two-condition construction is also possible: attend houses (ignore faces) and attend faces (ignore houses). There, each of the two conditions were presented 48 times across the four runs.
fMRI behavioral reponses
Participants’ behavioral responses during task fMRI were acquired via button boxes. This permitted examining performance during the fMRI tasks. After the second run of the IPT, data were also acquired regarding the participant’s perceived, overall monetary gains or losses during that task. Finally, at the end of each resting-state fMRI run, self-report data were acquired regarding whether participants had fallen asleep or whether they had trouble keeping their eyes open during that run.
Data and metadata were curated on the Extensible Neuroimaging Archive Toolkit (XNAT)37 server hosted at a study site. DICOM image files on the XNAT server were transmitted to the Connectome Coordination Facility. Quality control procedures and processing were completed within the Connectome Coordination Facility’s internal database28. Connectome Coordination Facility staff visually inspected unprocessed T1- and T2-weighted images for artifacts and other abnormalities that could interfere with analyses or interpretations. They provided quality ratings (4-point scale, poor – excellent) for these unprocessed images28. Unprocessed images underwent the HCP’s minimal preprocessing workflow38.
Minimally-preprocessed data
HCP minimal preprocessing workflows are detailed comprehensively elsewhere38,39 (humanconnectome.org/storage/app/media/documentation/s1200/HCP_S1200_Release_Reference_Manual.pdf#page = 125.19) However, a conceptual overview is provided in Fig. 2.
Conceptual overview and major steps of HCP’s minimal preprocessing workflows. Figure adapted from the HCP Young Adult article on minimal preprocessing workflows38.
Data Records
Data and metadata used here were curated using common structure definitions and data dictionaries where available within the NDA, as part of BANDA Release 1.140. This approach had the benefit of aggregating “shared subjects” using common data elements within a single repository, providing the end user with a capacity for efficient, cross-study queries. Participants are identifiable using a PseudoGUID (subjectkey), as well as a local ID (src_subject_id), which were mapped onto a common dictionary. Data dictionaries are available in delimited-text (.csv) format and may be accessed along with non-MRI and MRI data40. See Fig. 3 for an overview of data records.
Select data types, collections, and record structures. (a) Non-MRI (top) and MRI (bottom) data types. (b) MRI unprocessed superordinate directories (i.e., imagingcollection01), scan types, number of runs/directories, and some subdirectory information. (c) BANDA non-imaging data collections and associated NDA structure names. Note: K-SADS summary diagnoses provided are based upon adolescent report.
Imaging record structure
Unprocessed imaging collection
Figure 4 illustrates directory structures. Unprocessed NIFTI image files and associated metadata are curated within the NDA’s imagingcollection01 collection. The unprocessed collection includes button responses during task fMRI, post-scan responses to IPT and resting-state scan questions, and stimulus timing files for task fMRI runs in delimited-text formats (.csv and.txt). Text files containing scan information (e.g., date/time, flip angle, repetition time, b-values) were curated therein. Quality assurance scores for unprocessed images are available in session_report.csv files for each participant. On average, unprocessed data should require about 5-6 GB of local storage per participant directory (~1.3 TB total).
Minimally-preprocessed collection
Figure 5 illustrates examples of directory structures. Outputs from the HCP’s minimal preprocessing workflows were curated within the NDA’s fmriresults01 collection. This collection also provides associated workflow code, logs, and other environment information (e.g., QuNex outputs)41 used to generate the outputs. Quality control outputs provide snapshots (.png files) for efficient evaluation of select image types, as well as complete image maps (e.g.,.nii,.gii) that can be used for more comprehensive quality analyses. Figure 6 illustrates example outputs. Head-displacement estimates are also available for BOLD and diffusion-weighted images. For diffusion-weighted images, eddyqc outputs are provided that include quality control snapshots and documentation of signal quality, volume-to-volume motion/outlier qualification, and eddy currents via eddy_quad41. The BANDAAllFiles collection, containing these and unprocessed data, requires around ~5.9 TB of available local storage. The HCP also provides additional details on the standardized data structure (humanconnectome.org/storage/app/media/documentation/s1200/HCP_S1200_Release_Reference_Manual.pdf#page = 125.19).
fmriresults01 collection structure examples adapted from BANDA Release 1.1 Reference Manual Appendix. (a) Minimally preprocessed superordinate directories. (b) Subdirectories for diffusion-weighted outputs. (c) Subdirectories for T1-weighted outputs in native space. (d) Subdirectories for EPT outputs in standard spaces. (e) Subdirectories for T1-weighted outputs in standard spaces.
Example structural QC output adapted from fmriresults01 collection. Left: T1-weighted (top) and T2-weighted volumes in native space with pial (blue) and white matter (green) surfaces overlaid. Right: Inflated Conte69 cortical surface with unsmoothed myelin map (top) and midthickness native surface with curvature map (bottom). QC snapshot images were edited for space and facial features were obscured.
Structural paths
fmriresults01/ < subject_MR > /T1w/ and its subordinate directories include: T1w and T2w volumes; native Freesurfer surfaces/volumes; segmentation/parcellation outputs and statistics (e.g., aseg.stats); as well data in original acquisition spaces after AC-PC alignment. fmriresults01/ < subject_MR > /MNINonLinear/ includes: surfaces and other data nonlinearly registered to MNI152 space (and Conte69 surface); AC-PC transforms and native space to MNI152 transforms (i.e., /xfms/); as well as data resampled the to the fsaverage 32k mesh.
fMRI paths
fmriresults01/ < subject_MR > /MNINonLinear/Results/ includes: volume and grayordinate data for task and resting state data; 12 degrees-of-freedom motion parameters (i.e., Movement_Regressors.txt) among others; and task fMRI event times (i.e., EVs).
Diffusion paths
fmriresults01/ < subject_MR > /T1w/Diffusion/ and its subordinate directories include: b-value and b-vector files; a preprocessed diffusion 4-D file (data.nii.gz); a brain mask in diffusion space (nodif_brain_mask.nii.gz); and the estimated voxel-wise effects of gradient nonlinearities on b-values and b-vectors (grad_dev.nii.gz).
Technical Validation
Summary
This article provides technical validations newly performed on the completed BANDA sample (N = 215). Technical validation was demonstrated in three ways. First, the ndarsubject01 phenotype labels (i.e., Anxiety, Depression, and Control) were demonstrated to exhibit expected relationships with clinical and neurocognitive data obtained during Session 1. Second, the MRI-related quality metrics (e.g., signal quality measures, head displacement, button responses) were demonstrated to have no apparent relationships to the ndarsubject01 phenotype labels. Finally, the minimally-preprocessed imaging data were shown to be capable of producing canonical task-based and resting-fMRI signatures with limited additional processing. Demonstrations of preliminary validations performed on the initial 140 participants’ pre-release data are also available. Pre-release validations included inter-rater agreements of diagnostic classifications and internal consistency of self-report data21. These also included qualitative and quantitative harmonization comparisons with HCP-D data, as well as demonstrations of the preliminary relationships between clinical symptoms/diagnoses and MRI quality metrics13.
Expected phenotype and clinical/neurocognitive relationships
Anxious or depressed adolescents should be distinct from controls on measures that assess internalizing symptoms. However, marked differences were not expected among these groups on measures assessing externalizing symptoms or neurocognitive performance. Figure 7 illustrates uniform manifold approximation and projection embeddings (UMAP)42 recovered from internalizing, externalizing, and neurocognitive feature sets (see Table 1). In sum, Anxiety and Depression phenotypes, expectedly, exhibited apparent separation from Controls on dimensions derived from internalizing features. Also consistent with expectations, minimal separation was observed among phenotypes on externalizing dimensions and no apparent separation was observed among them on neurocognitive dimensions.
Phenotype labels projected onto two-dimensional representations of feature sets from select study elements. Dimensions extracted using UMAP for each feature set. Table 1 details elements used for feature sets.
Measures
Twenty-nine data elements were extracted from 16 NDA structures available. Table 1 describes measures used and the feature sets these were assigned to. All measures were taken from available Session 1 adolescent report and parent-report-on-adolescent data. The UMAP reduction was executed within the MATLAB2022b environment using run_umap program within the Uniform Manifold Approximation and Projection package (v.4.4)43. Default parameters were used, excluding user specification of the distance metric (‘seculidean’) and the neighborhood search method (‘exhaustive’).
Expected phenotype effects on MRI-related quality metrics
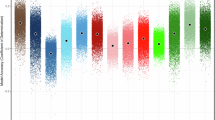
Figure 8 demonstrates phenotype effects on 33 metrics that could impact MRI quality or subsequent group comparisons. Bayesian analyses failed to show greater than anecdotal strength of evidence in favor of the alternative hypothesis of differences amongst phenotype labels on these metrics—with at most anecdotal evidence in favor of the alternative for only 3 of the 33 metrics. Conversely, 48% of analyses indicated moderate or strong evidence favoring the null hypothesis compared to a phenotype effect on MRI quality metrics. In sum, evidence failed to support that phenotypes differed on measures that could influence MRI quality. The evidence more broadly indicated support in favor of the null hypothesis—that phenotypes did not differ on MRI quality metrics.
Bayes factors for phenotype effects on MRI-related quality metrics. Colors indicate measure type, bar shade indicates qualification of the strength of evidence for null or alternative hypothesis. Anatomical signal quality (green); anatomical quality ratings (brown); diffusion signal quality (blue); frame-wise displacement during functional imaging (yellow); motivation ratings during functional imaging tasks (purple); performance on functional imaging tasks (orange).
T1- and T2-weighted metrics
AFNI was used for additional analyses of minimally preprocessed data44. Minimally preprocessed T1- and T2-weighted images were used (e.g., T1.nii.gz) along with participants’ gray- and white-matter segmentation maps (wmparc.nii.gz). Average gray matter signal intensity was extracted from a bilateral sample of cortical (medial prefrontal, superior parietal, fusiform) and subcortical regions (thalamus), as well as cerebellar cortex. Average white matter signal intensity was extracted from bilateral unsegmented white matter13, as well as bilateral cerebellar white matter. Background noise was estimated using the standard deviation of voxel intensities from a volume that did not contain human tissue—a 10 mm3 sphere placed in the right, anterior-superior corner of the image frame13. Contrast-to-noise ratios (CNR) were estimated by subtracting average gray- from white-matter signal intensity and dividing by the background noise for T1-weighted images. CNR was estimated similarly for T2-weighted images, but average white-matter signal intensity was instead subtracted from average gray-matter signal intensity. Signal-to-noise ratios (SNR) were estimated using the average of gray- and white-matter intensities divided by background noise. Quality ratings from visual inspections provided by Connectome Coordination Facility staff were also examined.
Diffusion metrics
Outputs from minimally preprocessed diffusion-weighted images were used. These were generated as part of the HCP’s eddyqc workflow (cnr.nii.gz)39 which provided SNR of b0, as well as CNR of b1500 and b3000 images. These metrics were extracted from the average of voxels contained within the white matter mask described above.
fMRI metrics
Participant motion was quantified from each functional run using their average framewise displacement (FD) calculated from the 6 degrees-of-freedom displacement values available in their minimally preprocessed outputs (Movement_Regressors.txt). Reports of participants’ motivation for performing well on each fMRI task were also collected using a rating scale of 1–10 (not motivated at all – very motivated). Ratings indicated that motivation was skewed toward performing well on each of the three tasks with modal ratings of 8 (Median = 7) for the EPT, 8 (Median = 7) for the IPT, and 7 (Median = 6) for the EIT. Ratings were collected as part of an internal scanning questionnaire used to monitor incoming task fMRI data and thus, were not part of the data release. While phenotypes were expected to differ on specific behavioral contrasts on the fMRI tasks (e.g., EIT ignore fearful faces > attend fearful faces)45, phenotypes were not expected to differ in their overall task performance. Thus, we also sought to examine whether differences existed among phenotype labels on overall fMRI task performance. These included average response accuracy computed from the EIT and EPT participants’ button-response outputs (e.g., < subject > _task-conflict_run-01.csv) and average response bias calculated from IPT outputs21. Average response time and the number of skipped responses were also evaluated.
Expected task and resting-state fMRI signatures
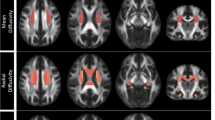
Figure 9 illustrates conservatively thresholded voxel clusters (p < 0.001; k = 100 faces-touching voxel clusters) for select task-fMRI contrasts using volume data from participants’ minimally-preprocessed fMRIVolume workflow outputs. The EPT’s Faces > Objects contrast elicited expected, suprathreshold activations in core and extended face-network regions, including bilateral amygdalae, fusiform gyri, and dorsolateral prefrontal regions (Fig. 9a)32,33,46. Relatedly, the EIT’s Attend Faces > Attend Houses contrast elicited expected, suprathreshold activations in bilateral amygdalae and fusiform gyri (Fig. 9b)34,36. The IPT’s Reward > Loss contrast elicited expected, suprathreshold activations primarily in bilateral ventral striatum (Fig. 9c)30,31,47. Figure 10 illustrates the top 95% of voxels with the greatest connectivity with a single, a priori determined precuneus voxel48. This map was derived from all available participants’ minimally-preprocessed outputs and demonstrates the expected recovery of canonical default-mode network areas, including precuneus/post-cingulate cortex, medial prefrontal cortex, as well as portions of bilateral parietal lobules and middle temporal gyri49,50.
Processing
The purpose of these analyses was to illustrate that expected task- and resting-fMRI signatures could be recovered, even with limited additional processing procedures. We encourage the use of more rigorous processing procedures for hypothesis testing (e.g., motion censoring, white matter/CSF regressions, age-/sex- regression). However, we chose to omit these procedures to demonstrate the findings that could be observed given only a limited set of common analytic decisions.
Task fMRI
Minimally preprocessed volume outputs (e.g., tfMRI_GAMBLING_AP.nii.gz) were spatially smoothed using a 6 mm Gaussian kernel constrained within their associated whole-brain mask (e.g., tfMRI_GAMBLING_AP_finalmask.nii.gz). Generalized linear models from AFNI’s 3dDeconvolve were used to estimate task activations by convolving image timeseries with canonical impulse response functions (i.e., block for EPT and IPT; double-gamma for EIT), stimulus timings extracted from EV design files (e.g., loss.txt, reward.txt), and controlling for the 12 degrees-of-freedom head displacement estimates (i.e., Movement_Regressors.txt) and other nuisance signals (e.g., signal drifts). Beta weights were averaged across participants’ runs, aggregated, and then subjected to paired-sample t-tests to produce voxel-wise, parametric maps of contrasts constrained within a group-level brain mask using AFNI’s 3dttest++ program.
Resting fMRI
Minimally preprocessed outputs (e.g., rfMRI_REST1_AP.nii.gz) were processed using AFNI’s 3dTproject program. The workflow employed bandpass filtering (0.01–0.1 Hz), controlled for 12 degrees-of-freedom head displacement estimates (i.e., Movement_Regressors.txt), and used 6 mm spatial smoothing (6 mm Gaussian kernel) constrained within participants’ associated whole-brain mask (e.g., rfMRI_REST1_AP_finalmask.nii.gz). Runs were then concatenated. Voxel-wise functional connectivity analyses were performed within a group-level brain mask using AFNI’s 3dGroupInCorr program with a single precuneus voxel (RAI: +01, +50, +28)48.
Usage Notes
Accessing data and recommended resources
BANDA data (Release Version 1.1)40 can be accessed via the nda.nih.gov website. Obtaining access to BANDA data requires an active NDA user account and a Data Use Certification. Acceptance of the NDA’s data-use terms and conditions is required. Notifications on future data releases or updates may be obtained by subscribing to the HCP’s listserv.
The BANDA Data Release 1.1 Reference Manual and Appendices contain further details regarding the study, specific study measures and NDA directory structures, as well as illustrated walkthroughs for creating and managing BANDA download packages from the NDA. These are accessible via the associated NDA repository40 or can be downloaded directly from the BANDA Resources and Materials repository51. The latter repository also contains documentation on missing data (e.g., BANDA1.1_Completeness.csv), data dictionaries (BANDA1.1_Crosswalk.csv), and the specific imaging protocols used to obtain BANDA MRI data.
Critical considerations for fMRI analyses
Excluded frames
As part of the Connectome Coordination Facility’s workflow, processed and unprocessed data released to the NDA excluded the first 10 frames of each resting state, IPT, and EIT runs. For these tasks, stimulus design files (i.e., EVs) released to the NDA were adjusted to compensate for these excluded frames. The excluded frames are available as part of the imagingcollection01 structure (< subject_MR >/unprocessed/OTHER_FILES/*_ InitialFrames.nii.gz). However, if these excluded frames are used for task-based analyses, the stimulus design files must be adjusted accordingly.
EPT design
The sad image condition was added after the first 16 participants thus, BANDA001-BANDA016 have fewer frames and different stimulus design files (i.e., EVs) compared to BANDA017-BANDA215. Additionally, the stimulus design files released also include fixation.txt files. These do not reflect an “active” task condition and will not be necessary for most use cases.
Critical considerations for Non-MRI data analyses
Session descriptions vs. NDA visit element
The data acquisition sessions described here (i.e., Sessions 1–4) should not be conflated with the NDA visit element (T1–T4)40. Therein, visit T1 refers to data collected during the initial intake or brain imaging visit, T2 refers to data collected during the 6-month follow-up visit, and T3 refers to data collected during the 12-month follow-up visit. Data for a visit T4 is also available from a small subset of participants (n = 22) who were recontacted after T3 as part of a separate, pilot study. However, because visit T4 data were not collected using the exact procedures as visits T1–T3, it is not recommended that prospective users incorporate these data into longitudinal analyses.
Referencing BANDA
To allow consistent tracking of BANDA-based research products, we recommend that researchers using BANDA data include the following text in their published works in addition to referencing this publication: “Data were provided [in part] by the Boston Adolescent Neuroimaging of Anxiety and Depression (BANDA) Consortium’s Human Connectome Project, supported by 1U01MH108168 (PIs: Susan Whitfield-Gabrieli, John Gabrieli).”51 Future studies using BANDA imaging data should cite Siless and colleagues13, which provides details on imaging protocols, hardware, and harmonization with other HCP studies. Future studies using functional imaging tasks, clinical or cognitive data should cite Hubbard and colleagues21. The fMRI tasks were adapted from original works (IPT30; EPT32,33; EIT34,35,36). Researchers describing BANDA fMRI tasks are encouraged to also refer to those original works.
Code availability
Code for transforming select item-level, raw data to NDA summary/composite score elements is available within the BANDA Resources and Materials repository51. Source code for the fMRI tasks is accessible via the BANDA fMRI-tasks repository52. Code used to generate the quality assurance and HCP minimally-preprocessed outputs is curated within the BANDAAllFiles collection NDA package34 (e.g., /fmriresults01/< subject_MR >/ProcessingInfo).
References
Casey, B. J. Beyond simple models of self-control to circuit-based accounts of adolescent behavior. Annu. Rev. Psychol. 66, 295–319 (2015).
Somerville, L. H., Jones, R. M. & Casey, B. A time of change: Behavioral and neural correlates of adolescent sensitivity to appetitive and aversive environmental cues. Brain Cogn 72, 124 (2010).
Auerbach, R. P., Webb, C. A., Gardiner, C. K. & Pechtel, P. Behavioral and neural mechanisms underlying cognitive vulnerability models of depression. Journal of Psychotherapy Integration 23, 222–235 (2013).
Hofmann, S. G., Ellard, K. K. & Siegle, G. J. Neurobiological correlates of cognitions in fear and anxiety: A cognitive–neurobiological information-processing model. Cognition & Emotion 26, 282–299 (2012).
Pizzagalli, D. A. Depression, stress, and anhedonia: Toward a synthesis and integrated model. Annu Rev Clin Psychol 10, 393–423 (2014).
Volkow, N. D. et al. The conception of the ABCD study: From substance use to a broad NIH collaboration. Developmental Cognitive Neuroscience 32, 4–7 (2018).
Schumann, G. et al. The imagen study: Reinforcement-related behaviour in normal brain function and psychopathology. Molecular Psychiatry 15, 1128–1139 (2010).
Satterthwaite, T. D. et al. The Philadelphia neurodevelopmental cohort: A publicly available resource for the study of normal and abnormal brain development in youth. NeuroImage 124, 1115–1119 (2016).
Harms, M. P. et al. Extending the human connectome project across ages: Imaging protocols for the lifespan development and aging projects. NeuroImage 183, 972–984 (2018).
Somerville, L. H. et al. The lifespan human connectome project in development: A large-scale study of brain connectivity development in 5–21 year olds. NeuroImage 183, 456–468 (2018).
Elam, J. S. et al. The human connectome project: A retrospective. NeuroImage 244, 118543 (2021).
Glasser, M. F. et al. The human connectome project’s neuroimaging approach. Nat Neurosci 19, 1175–1187 (2016).
Siless, V. et al. Image acquisition and quality assurance in the Boston adolescent neuroimaging of depression and anxiety study. NeuroImage: Clinical 26, 102242 (2020).
Kaufman, J. et al. Schedule for affective disorders and schizophrenia for school-age children-present and lifetime version (K-SADS-PL): Initial reliability and validity data. Journal of the American Academy of Child & Adolescent Psychiatry 36, 980–988 (1997).
American Psychiatric Association. Diagnostic and statistical manual of mental disorders, https://doi.org/10.1176/appi.books.9780890425596 (2013).
Weissman, M. M. et al. Brief screening for family psychiatric history: The family history screen. Archives of General Psychiatry 57, 675–682 (2000).
Posner, K. et al. The Columbia–suicide severity rating scale: Initial validity and internal consistency findings from three multisite studies with adolescents and adults. Am J Psychiatry 168, 1266–1277 (2011).
Wechsler, D. Wechsler Abbreviated Scale of Intelligence, Second Edition, https://doi.org/10.1037/t15171-000 (2011).
Harris, P. A. et al. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. Journal of Biomedical Informatics 42, 377–381 (2009).
Freedman, R. et al. The initial field trials of DSM-5: New blooms and old thorns. American Journal of Psychiatry 170, 1–5 (2013).
Hubbard, N. A. et al. Brain function and clinical characterization in the Boston adolescent neuroimaging of depression and anxiety study. NeuroImage: Clinical 27, 102240 (2020).
Taylor, S. J. et al. Performance of a new pubertal self‐assessment questionnaire: A preliminary study. Paediatric and Perinatal Epidemiology 15, 88–94 (2001).
Chapman, L. J. & Chapman, J. P. The measurement of handedness. Brain and Cognition 6, 175–183 (1987).
Slavich, G. M., Stewart, J. G., Esposito, E. C., Shields, G. S. & Auerbach, R. P. The stress and adversity inventory for adolescents (adolescent STRAIN): Associations with mental and physical health, risky behaviors, and psychiatric diagnoses in youth seeking treatment. Child Psychology Psychiatry 60, 998–1009 (2019).
Gershon, R. C. et al. NIH toolbox for assessment of neurological and behavioral function. Neurology 80 (2013).
Heaton, R. K. et al. Reliability and validity of composite scores from the NIH toolbox cognition battery in adults. J Int Neuropsychol Soc 20, 588–598 (2014).
Gur, R. C. et al. A cognitive neuroscience-based computerized battery for efficient measurement of individual differences: Standardization and initial construct validation. Journal of Neuroscience Methods 187, 254–262 (2010).
Marcus, D. S. et al. Human connectome project informatics: Quality control, database services, and data visualization. NeuroImage 80, 202–219 (2013).
Tisdall, M. D. et al. Volumetric navigators for prospective motion correction and selective reacquisition in neuroanatomical MRI. Magnetic Resonance in Med 68, 389–399 (2012).
Delgado, M. R., Nystrom, L. E., Fissell, C., Noll, D. C. & Fiez, J. A. Tracking the hemodynamic responses to reward and punishment in the striatum. Journal of Neurophysiology 84, 3072–3077 (2000).
Barch, D. M. et al. Function in the human connectome: Task-FMRI and individual differences in behavior. NeuroImage 80, 169–189 (2013).
Chai, X. J. et al. Functional and structural brain correlates of risk for major depression in children with familial depression. NeuroImage: Clinical 8, 398–407 (2015).
Hariri, A. R., Tessitore, A., Mattay, V. S., Fera, F. & Weinberger, D. R. The amygdala response to emotional stimuli: A comparison of faces and scenes. NeuroImage 17, 317–323 (2002).
Fales, C. L. et al. Altered emotional interference processing in affective and cognitive-control brain circuitry in major depression. Biological Psychiatry 63, 377–384 (2008).
Vuilleumier, P., Armony, J. L., Driver, J. & Dolan, R. J. Effects of attention and emotion on face processing in the human brain. Neuron 30, 829–841 (2001).
Wojciulik, E., Kanwisher, N. & Driver, J. Covert visual attention modulates face-specific activity in the human fusiform gyrus: Fmri study. Journal of Neurophysiology 79, 1574–1578 (1998).
Marcus, D. S., Olsen, T. R., Ramaratnam, M. & Buckner, R. L. The extensible neuroimaging archive toolkit: An informatics platform for managing, exploring, and sharing neuroimaging data. Neuroinformatics 5, 11–33 (2007).
Glasser, M. F. et al. The minimal preprocessing pipelines for the human connectome project. Neuroimage 80, 105–124 (2013).
Bastiani, M. et al. Automated quality control for within and between studies diffusion MRI data using a non-parametric framework for movement and distortion correction. NeuroImage 184, 801–812 (2019).
Elam, J. et al. Data repository for: Boston adolescent neuroimaging of depression & anxiety (BANDA) release 1.1. NIMH Data Repositories https://doi.org/10.15154/3TK5-PB47 (2024).
Ji, J. L. et al. QuNex—an integrative platform for reproducible neuroimaging analytics. Frontiers in Neuroinformatics 17, (2023).
McInnes, L., Healy, J. & Melville, J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. Preprint at http://arxiv.org/abs/1802.03426 (2020).
Meehan, C., Ebrahimian, J., Moor, W. & Meehan, S. Uniform manifold approximation and projection (UMAP). MathWorks File Exchange Available at: https://www.mathworks.com/matlabcentral/fileexchange/71902-uniform-manifold-approximation-and-projection-umap. (Last Accessed: 6th April 2024) (2022).
Cox, R. W. AFNI: Software for analysis and visualization of functional magnetic resonance neuroimages. Computers and Biomedical Research 29, 162–173 (1996).
Hubbard, N. A. et al. Connectivity patterns evoked by fearful faces demonstrate reduced flexibility across a shared dimension of adolescent anxiety and depression. Clinical Psychological Science 11, 3–22 (2023).
Haxby, J. V., Hoffman, E. A. & Gobbini, M. I. The distributed human neural system for face perception. Trends in Cognitive Sciences 4, 223–233 (2000).
Hubbard, N. A. et al. Reward-sensitive basal ganglia stabilize the maintenance of goal-relevant neural patterns in adolescents. Journal of Cognitive Neuroscience 32, 1508–1524 (2020).
Hubbard, N. A. et al. Resting cerebral oxygen metabolism exhibits archetypal network features. Human Brain Mapping 42, 1952–1968 (2021).
Whitfield-Gabrieli, S. & Ford, J. M. Default mode network activity and connectivity in psychopathology. Annu. Rev. Clin. Psychol. 8, 49–76 (2012).
Raichle, M. E. Two views of brain function. Trends in Cognitive Sciences 14, 180–190 (2010).
Hubbard, N. A., Bauer, C. C. C., Siless, V., Elam, J., & Ghosh, S. S. Resource repository for: BANDA resources and materials data release v1.1 (BANDA_v1.1_data_release). Zenodo https://doi.org/10.5281/zenodo.10849500 (2024).
Siless, V. & Bauer, C. C. C. Source code repository for: BANDA fMRI task code. Zenodo https://doi.org/10.5281/zenodo.10851055 (2024).
Auerbach, R. P. et al. Reward-related neural circuitry in depressed and anxious adolescents: A Human Connectome Project. J Amer Academy of Child and Adol Psychi 61, 308–320 (2022).
Acknowledgements
This project was supported by the National Institute of Mental Health, U01MH108168 (JDEG, SWG), and (F32MH114525 to NAH). NAH was partially supported by the Brain and Behavior Research Foundation (#27970) and the NIGMS (P20GM130461-6206). AY was partially supported by R01EB021265 and U01EB026996. DAP was partially supported by R37MH068376 and R01MH101521. SSG was partially supported by R01EB020740 and P41EB019936. RPA was partially supported by R01MH135488, R01MH119771. SGH was partly supported by R01AT007257, R01MH099021, the James S. McDonnell Foundation, the Alexander von Humboldt Foundation (Alexander von Humboldt Professur), and the Hessische Ministerium für Wissenschaft und Kunst (LOEWE Spitzenprofessur). This project was made possible by the resources provided by Shared Instrumentation Grants 1S10RR023401, 1S10RR019307, and 1S10RR023043. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or of any other sponsor.
Author information
Authors and Affiliations
Contributions
Hubbard: conceptualization, writing, visualization, validation, supervision, analysis, methodology, project administration; Bauer: data curation, validation, methodology, analysis; Siless: data curation, project administration, data collection, supervision, methodology; Auerbach: conceptualization, writing, validation, funding acquisition, project administration; Elam: analysis, data curation, methodology; Frosch: data collection, data curation, project administration; Henin: conceptualization, writing, data collection, supervision, funding acquisition, project administration; Hodge: data curation, methodology; Hofmann: conceptualization, writing, supervision, funding acquisition, project administration; Jones: data collection, data curation; Lenzini: analysis, data curation; Lo: analysis, data curation; Park: data curation, methodology; Pizzagalli: conceptualization, writing, supervision, funding acquisition, project administration; Vaz De-Souza: data collection, data curation; Gabrieli: conceptualization, writing, project administration, supervision, funding acquisition; Whitfield-Gabrieli: conceptualization, writing, project administration, supervision, funding acquisition; Yendiki: conceptualization, writing, project administration, data collection, supervision, methodology, funding acquisition; Ghosh: conceptualization, methodology, validation, data curation, writing, supervision, funding acquisition, project administration.
Corresponding authors
Ethics declarations
Competing interests
Over the past 3 years, Dr. Pizzagalli has received consulting fees from Albright Stonebridge Group, Boehringer Ingelheim, Compass Pathways, Engrail Therapeutics, Neumora Therapeutics (formerly BlackThorn Therapeutics), Neurocrine Biosciences, Neuroscience Software, Otsuka, Sunovion, and Takeda; he has received honoraria from the Psychonomic Society and American Psychological Association (for editorial work) and from Alkermes; he has received research funding from the Brain and Behavior Research Foundation, Dana Foundation, Wellcome Leap, Millennium Pharmaceuticals, and NIMH; he has received stock options from Compass Pathways, Engrail Therapeutics, Neumora Therapeutics, and Neuroscience Software. Dr. Auerbach is a paid scientific advisor for Get Sonar, Inc and an unpaid advisor for Ksana Health. Dr. Hofmann receives financial support by the Alexander von Humboldt Foundation (as part of the Alexander von Humboldt Professur), the Hessische Ministerium für Wissenschaft und Kunst (as part of the LOEWE Spitzenprofessur), and the DYNAMIC center, funded by the LOEWE program of the Hessian Ministry of Science and Arts (Grant Number: LOEWE1/16/519/03/09.001(0009)/98). He also receives compensation for his work as editor from SpringerNature and royalties and payments for his work from various publishers.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hubbard, N.A., Bauer, C.C.C., Siless, V. et al. The Human Connectome Project of adolescent anxiety and depression dataset. Sci Data 11, 837 (2024). https://doi.org/10.1038/s41597-024-03629-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-024-03629-x
- Springer Nature Limited